Overlapping Patterns of Gene Expression Between Gametophyte and Sporophyte Phases in the Fern Polypodium amorphum (Polypodiales)
- PMID: 30356815
- PMCID: PMC6190754
- DOI: 10.3389/fpls.2018.01450
Overlapping Patterns of Gene Expression Between Gametophyte and Sporophyte Phases in the Fern Polypodium amorphum (Polypodiales)
Abstract
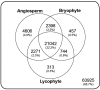
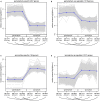
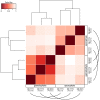
Ferns are unique among land plants in having sporophyte and gametophyte phases that are both free living and fully independent. Here, we examine patterns of sporophytic and gametophytic gene expression in the fern Polypodium amorphum, a member of the homosporous polypod lineage that comprises 80% of extant fern diversity, to assess how expression of a common genome is partitioned between two morphologically, ecologically, and nutritionally independent phases. Using RNA-sequencing, we generated transcriptome profiles for three replicates of paired samples of sporophyte leaf tissue and whole gametophytes to identify genes with significant differences in expression between the two phases. We found a nearly 90% overlap in the identity and expression levels of the genes expressed in both sporophytes and gametophytes, with less than 3% of genes uniquely expressed in either phase. We compare our results to those from similar studies to establish how phase-specific gene expression varies among major land plant lineages. Notably, despite having greater similarity in the identity of gene families shared between P. amorphum and angiosperms, P. amorphum has phase-specific gene expression profiles that are more like bryophytes and lycophytes than seed plants. Our findings suggest that shared patterns of phase-specific gene expression among seed-free plants likely reflect having relatively large, photosynthetic gametophytes (compared to the gametophytes of seed plants that are highly reduced). Phylogenetic analyses were used to further investigate the evolution of phase-specific expression for the phototropin, terpene synthase, and MADS-box gene families.
Keywords: MADS-box; gametophyte; life cycle; phototropins; sporophyte; terpene synthases; transcriptomics.
Figures



References
-
- Ambrose B. (2010). MADS-box genes in plant evolution and development. Int J Plant Dev. Biol. 4 30–37.
-
- Andrews S. (2010). FastQC: a Quality Control Tool for High Throughput Sequence Data. Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc
-
- Barker M. S., Wolf P. G. (2010). Unfurling fern biology in the age of genomics. Bioscience 60 177–185. 10.1525/bio.2010.60.3.4 - DOI
Associated data
LinkOut - more resources
Full Text Sources

