BitterDB: taste ligands and receptors database in 2019
- PMID: 30357384
- PMCID: PMC6323989
- DOI: 10.1093/nar/gky974
BitterDB: taste ligands and receptors database in 2019
Abstract
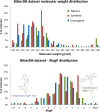
BitterDB (http://bitterdb.agri.huji.ac.il) was introduced in 2012 as a central resource for information on bitter-tasting molecules and their receptors. The information in BitterDB is frequently used for choosing suitable ligands for experimental studies, for developing bitterness predictors, for analysis of receptors promiscuity and more. Here, we describe a major upgrade of the database, including significant increase in content as well as new features. BitterDB now holds over 1000 bitter molecules, up from the initial 550. When available, quantitative sensory data on bitterness intensity as well as toxicity information were added. For 270 molecules, at least one associated bitter taste receptor (T2R) is reported. The overall number of ligand-T2R associations is now close to 800. BitterDB was extended to several species: in addition to human, it now holds information on mouse, cat and chicken T2Rs, and the compounds that activate them. BitterDB now provides a unique platform for structure-based studies with high-quality homology models, known ligands, and for the human receptors also data from mutagenesis experiments, information on frequently occurring single nucleotide polymorphisms and links to expression levels in different tissues.
Figures
Similar articles
-
BitterDB: 2024 update on bitter ligands and taste receptors.Nucleic Acids Res. 2025 Jan 6;53(D1):D1645-D1650. doi: 10.1093/nar/gkae1044. Nucleic Acids Res. 2025. PMID: 39535052 Free PMC article.
-
BitterDB: a database of bitter compounds.Nucleic Acids Res. 2012 Jan;40(Database issue):D413-9. doi: 10.1093/nar/gkr755. Epub 2011 Sep 22. Nucleic Acids Res. 2012. PMID: 21940398 Free PMC article.
-
Bitterness prediction in-silico: A step towards better drugs.Int J Pharm. 2018 Feb 5;536(2):526-529. doi: 10.1016/j.ijpharm.2017.03.076. Epub 2017 Mar 28. Int J Pharm. 2018. PMID: 28363856
-
Bitter taste receptors: Novel insights into the biochemistry and pharmacology.Int J Biochem Cell Biol. 2016 Aug;77(Pt B):184-96. doi: 10.1016/j.biocel.2016.03.005. Epub 2016 Mar 16. Int J Biochem Cell Biol. 2016. PMID: 26995065 Review.
-
Recent advances in structure and function studies on human bitter taste receptors.Curr Protein Pept Sci. 2012 Sep;13(6):501-8. doi: 10.2174/138920312803582942. Curr Protein Pept Sci. 2012. PMID: 23061795 Review.
Cited by
-
Rational design of agonists for bitter taste receptor TAS2R14: from modeling to bench and back.Cell Mol Life Sci. 2020 Feb;77(3):531-542. doi: 10.1007/s00018-019-03194-2. Epub 2019 Jun 24. Cell Mol Life Sci. 2020. PMID: 31236627 Free PMC article.
-
Inhibiting a promiscuous GPCR: iterative discovery of bitter taste receptor ligands.Cell Mol Life Sci. 2023 Apr 3;80(4):114. doi: 10.1007/s00018-023-04765-0. Cell Mol Life Sci. 2023. PMID: 37012410 Free PMC article.
-
VirtualTaste: a web server for the prediction of organoleptic properties of chemical compounds.Nucleic Acids Res. 2021 Jul 2;49(W1):W679-W684. doi: 10.1093/nar/gkab292. Nucleic Acids Res. 2021. PMID: 33905509 Free PMC article.
-
Bitter taste sensitivity in domestic dogs (Canis familiaris) and its relevance to bitter deterrents of ingestion.PLoS One. 2022 Nov 30;17(11):e0277607. doi: 10.1371/journal.pone.0277607. eCollection 2022. PLoS One. 2022. PMID: 36449493 Free PMC article.
-
Taste Sensor Assessment of Bitterness in Medicines: Overview and Recent Topics.Sensors (Basel). 2024 Jul 24;24(15):4799. doi: 10.3390/s24154799. Sensors (Basel). 2024. PMID: 39123846 Free PMC article. Review.
References
-
- Chandrashekar J., Hoon M.A., Ryba N.J.P., Zuker C.S.. The receptors and cells for mammalian taste. Nature. 2006; 444:288–294. - PubMed
-
- Behrens M., Korsching S.I., Meyerhof W.. Tuning properties of avian and frog bitter taste receptors dynamically fit gene repertoire sizes. Mol. Biol. Evol. 2014; 31:3216–3227. - PubMed
-
- Behrens M., Meyerhof W.. Vertebrate bitter taste receptors: keys for survival in changing environments. J. Agric. Food Chem. 2018; 66:2204–2213. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

