Gene Coexpression Networks in Whole Blood Implicate Multiple Interrelated Molecular Pathways in Obesity in People with Asthma
- PMID: 30358166
- PMCID: PMC6262830
- DOI: 10.1002/oby.22341
Gene Coexpression Networks in Whole Blood Implicate Multiple Interrelated Molecular Pathways in Obesity in People with Asthma
Abstract
Objective: Asthmatic children who develop obesity through adolescence have poorer disease outcomes compared with those who do not. This study aimed to characterize the biology of childhood asthma complicated by adult obesity.
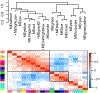
Methods: Gene expression networks are powerful statistical tools for characterizing human disease that leverage the putative coregulatory relationships of genes to infer relevant biological pathways. Weighted gene coexpression network analysis of gene expression data was performed in whole blood from 514 adult asthmatic subjects. Then, module preservation and association replication analyses were performed in 418 subjects from two independent asthma cohorts (one pediatric and one adult).
Results: A multivariate model was identified in which three gene coexpression network modules were associated with incident obesity in the discovery cohort (each P < 0.05). Two module memberships were enriched for genes in pathways related to platelets, integrins, extracellular matrix, smooth muscle, NF-κB signaling, and Hedgehog signaling. The network structures of each of the obesity modules were significantly preserved in both replication cohorts (permutation P = 9.999E-05). The corresponding module gene sets were significantly enriched for differential expression in subjects with obesity in both replication cohorts (each P < 0.05).
Conclusions: The gene coexpression network profiles thus implicate multiple interrelated pathways in the biology of an important endotype of asthma with obesity.
© 2018 The Obesity Society.
Figures



References
-
- Gibson PG. Obesity and asthma. Ann Am Thorac Soc 2013;10 Suppl: S138–142. - PubMed
-
- Scott HA, Wood LG, Gibson PG. What About Neutrophils in Obese Asthma? Am J Respir Cell Mol Biol 2016;55: 462. - PubMed
-
- Gruchala-Niedoszytko M, Niedoszytko M, Sanjabi B, van der Vlies P, Niedoszytko P, Jassem E, et al. Analysis of the differences in whole-genome expression related to asthma and obesity. Pol Arch Med Wewn 2015;125: 722–730. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

