Transcriptome Analysis Reveals Nonfoamy Rather Than Foamy Plaque Macrophages Are Proinflammatory in Atherosclerotic Murine Models
- PMID: 30359200
- PMCID: PMC6945121
- DOI: 10.1161/CIRCRESAHA.118.312804
Transcriptome Analysis Reveals Nonfoamy Rather Than Foamy Plaque Macrophages Are Proinflammatory in Atherosclerotic Murine Models
Abstract
Rationale: Monocyte infiltration into the subintimal space and its intracellular lipid accumulation are the most prominent features of atherosclerosis. To understand the pathophysiology of atherosclerotic disease, we need to understand the characteristics of lipid-laden foamy macrophages in the subintimal space during atherosclerosis.
Objective: We sought to examine the transcriptomic profiles of foamy and nonfoamy macrophages isolated from atherosclerotic intima.
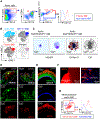
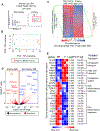
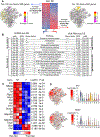
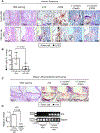
Methods and results: Single-cell RNA sequencing analysis of CD45+ leukocytes from murine atherosclerotic aorta revealed that there are macrophage subpopulations with distinct differentially expressed genes involved in various functional pathways. To specifically characterize the intimal foamy macrophages of plaque, we developed a lipid staining-based flow cytometric method for analyzing the lipid-laden foam cells of atherosclerotic aortas. We used the fluorescent lipid probe BODIPY493/503 and assessed side-scattered light as an indication of cellular granularity. BODIPYhiSSChi foamy macrophages were found residing in intima and expressing CD11c. Foamy macrophage accumulation determined by flow cytometry was positively correlated with the severity of atherosclerosis. Bulk RNA sequencing analysis showed that compared with nonfoamy macrophages, foamy macrophages expressed few inflammatory genes but many lipid-processing genes. Intimal nonfoamy macrophages formed the major population expressing IL (interleukin)-1β and many other inflammatory transcripts in atherosclerotic aorta.
Conclusions: RNA sequencing analysis of intimal macrophages from atherosclerotic aorta revealed that lipid-loaded plaque macrophages are not likely the plaque macrophages that drive lesional inflammation.
Keywords: RNA-seq; atherosclerosis; flow cytometry; foam cells; macrophages; mice.
Conflict of interest statement
DISCLOSURES
The authors declare no competing financial interests
Figures




Comment in
-
Who Done It? Macrophage Mayhem in Atherosclerosis.Circ Res. 2018 Oct 26;123(10):1106-1108. doi: 10.1161/CIRCRESAHA.118.314006. Circ Res. 2018. PMID: 30359193 Free PMC article. No abstract available.
-
Response by Kim and Choi to Letter Regarding Article, "Transcriptome Analysis Reveals Nonfoamy Rather Than Foamy Plaque Macrophages Are Proinflammatory in Atherosclerotic Murine Models".Circ Res. 2018 Nov 9;123(11):e50. doi: 10.1161/CIRCRESAHA.118.314163. Circ Res. 2018. PMID: 30571469 No abstract available.
-
Letter by Cochain et al Regarding Article, "Transcriptome Analysis Reveals Nonfoamy Rather Than Foamy Plaque Macrophages Are Proinflammatory in Atherosclerotic Murine Models".Circ Res. 2018 Nov 9;123(11):e48-e49. doi: 10.1161/CIRCRESAHA.118.314120. Circ Res. 2018. PMID: 30571470 No abstract available.
References
-
- Hansson GK, Hermansson A. The immune system in atherosclerosis. Nat Immunol. 2011;12:204–212 - PubMed
-
- Hansson GK, Libby P. The immune response in atherosclerosis: A double-edged sword. Nat Rev Immunol. 2006;6:508–519 - PubMed
-
- Weber C, Noels H. Atherosclerosis: Current pathogenesis and therapeutic options. Nat Med. 2011;17:1410–1422 - PubMed
-
- Suzuki H, Kurihara Y, Takeya M, Kamada N, Kataoka M, Jishage K, Ueda O, Sakaguchi H, Higashi T, Suzuki T, Takashima Y, Kawabe Y, Cynshi O, Wada Y, Honda M, Kurihara H, Aburatani H, Doi T, Matsumoto A, Azuma S, Noda T, Toyoda Y, Itakura H, Yazaki Y, Kodama T, et al. A role for macrophage scavenger receptors in atherosclerosis and susceptibility to infection. Nature. 1997;386:292–296 - PubMed
-
- Feig JE, Pineda-Torra I, Sanson M, Bradley MN, Vengrenyuk Y, Bogunovic D, Gautier EL, Rubinstein D, Hong C, Liu J, Wu C, van Rooijen N, Bhardwaj N, Garabedian M, Tontonoz P, Fisher EA. Lxr promotes the maximal egress of monocyte-derived cells from mouse aortic plaques during atherosclerosis regression. The Journal of clinical investigation. 2010;120:4415–4424 - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

