Clust: automatic extraction of optimal co-expressed gene clusters from gene expression data
- PMID: 30359297
- PMCID: PMC6203272
- DOI: 10.1186/s13059-018-1536-8
Clust: automatic extraction of optimal co-expressed gene clusters from gene expression data
Abstract
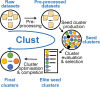
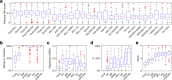
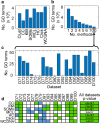
Identifying co-expressed gene clusters can provide evidence for genetic or physical interactions. Thus, co-expression clustering is a routine step in large-scale analyses of gene expression data. We show that commonly used clustering methods produce results that substantially disagree and that do not match the biological expectations of co-expressed gene clusters. We present clust, a method that solves these problems by extracting clusters matching the biological expectations of co-expressed genes and outperforms widely used methods. Additionally, clust can simultaneously cluster multiple datasets, enabling users to leverage the large quantity of public expression data for novel comparative analysis. Clust is available at https://github.com/BaselAbujamous/clust .
Keywords: Click; Clust; Clustering; Cross-clustering; Gene expression data; Hierarchical clustering; K-means; Markov clustering; Self-organizing maps; WGCNA.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

