Dissociation of Rpb4 from RNA polymerase II is important for yeast functionality
- PMID: 30359412
- PMCID: PMC6201915
- DOI: 10.1371/journal.pone.0206161
Dissociation of Rpb4 from RNA polymerase II is important for yeast functionality
Abstract
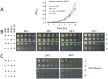
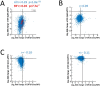
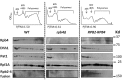
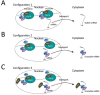
Rpb4 is an RNA polymerase II (Pol II) subunit that binds Pol II transcripts co-transcriptionally, accompanies them to the cytoplasm and modulates mRNA export, translation and decay by interacting with cytoplasmic RNA modulators. The importance of the cytoplasmic roles of Rpb4 was challenged by a study reporting that the phenotype of rpb2Δ rpb4Δ cells can be rescued by an Rpb2-Rpb4 fusion protein, assuming that its Rpb4 moiety cannot dissociate from Pol II and functions in the cytoplasm. Here we demonstrate that although the fusion protein supports normal transcription, it adversely affects mRNA decay, cell proliferation and adaptability-e.g., response to stress. These defects are similar, albeit milder, than the defects that characterize rpb4Δ cells. At least two mechanisms alleviate the deleterious effect of the fusion protein. First, a portion of this fusion protein is cleaved into free Rpb2 and Rpb4. The free Rpb4 is functional, as it binds mRNAs and polysomes, like WT Rpb4. Second, the fusion protein is also capable of binding poly(A)+ mRNAs in the cytoplasm, in an Rpb7-mediated manner, probably complementing the functions of the diminished Rpb4. Collectively, normal coupling between mRNA synthesis and decay requires wild-type configuration of Rpb4, and fusing Rpb4 to Rpb2 compromises this coupling.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

