Structural and Functional Analysis of a Bidirectional Promoter from Gossypium hirsutum in Arabidopsis
- PMID: 30360512
- PMCID: PMC6274729
- DOI: 10.3390/ijms19113291
Structural and Functional Analysis of a Bidirectional Promoter from Gossypium hirsutum in Arabidopsis
Abstract
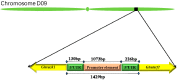
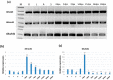
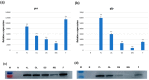
Stacked traits have become an important trend in the current development of genomically modified crops. The bidirectional promoter can not only prevent the co-suppression of multigene expression, but also increase the efficiency of the cultivation of transgenic plants with multigenes. In Gossypium hirsutum, Ghrack1 and Ghuhrf1 are head-to-head gene pairs located on chromosome D09. We cloned the 1429-bp intergenic region between the Ghrack1 and Ghuhrf1 genes from Gossypium hirsutum. The cloned DNA fragment GhZU had the characteristics of a bidirectional promoter, with 38.7% G+C content, three CpG islands and no TATA-box. Using gfp and gus as reporter genes, a series of expression vectors were constructed into young leaves of tobacco. The histochemical GUS (Beta-glucuronidase) assay and GFP (green fluorescence protein) detection results indicated that GhZU could drive the expression of the reporter genes gus and gfp simultaneously in both orientations. Furthermore, we transformed the expression vectors into Arabidopsis and found that GUS was concentrated at vigorous growth sites, such as the leaf tip, the base of the leaves and pod, and the stigma. GFP was also mainly expressed in the epidermis of young leaves. In summary, we determined that the intergenic region GhZU was an orientation-dependent bidirectional promoter, and this is the first report on the bidirectional promoter from Gossypium hirsutum. Our findings in this study are likely to enhance understanding on the regulatory mechanisms of plant bidirectional promoters.
Keywords: Gossypium hirsutum; bidirectional promoter; cloning; stable expression; transient expression.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
An intergenic region shared by At4g35985 and At4g35987 in Arabidopsis thaliana is a tissue specific and stress inducible bidirectional promoter analyzed in transgenic arabidopsis and tobacco plants.PLoS One. 2013 Nov 19;8(11):e79622. doi: 10.1371/journal.pone.0079622. eCollection 2013. PLoS One. 2013. PMID: 24260266 Free PMC article.
-
Isolation and Functional Characterization of a Constitutive Promoter in Upland Cotton (Gossypium hirsutum L.).Int J Mol Sci. 2024 Feb 5;25(3):1917. doi: 10.3390/ijms25031917. Int J Mol Sci. 2024. PMID: 38339199 Free PMC article.
-
A Gossypium hirsutum GDSL lipase/hydrolase gene (GhGLIP) appears to be involved in promoting seed growth in Arabidopsis.PLoS One. 2018 Apr 5;13(4):e0195556. doi: 10.1371/journal.pone.0195556. eCollection 2018. PLoS One. 2018. PMID: 29621331 Free PMC article.
-
Cloning and Functional Analysis of the Promoter of an Ascorbate Oxidase Gene from Gossypium hirsutum.PLoS One. 2016 Sep 6;11(9):e0161695. doi: 10.1371/journal.pone.0161695. eCollection 2016. PLoS One. 2016. PMID: 27597995 Free PMC article.
-
Promoter of a cotton fibre MYB gene functional in trichomes of Arabidopsis and glandular trichomes of tobacco.J Exp Bot. 2008;59(13):3533-42. doi: 10.1093/jxb/ern204. Epub 2008 Aug 18. J Exp Bot. 2008. PMID: 18711121 Free PMC article.
Cited by
-
Plant Defenses Against Pests Driven by a Bidirectional Promoter.Front Plant Sci. 2019 Jul 17;10:930. doi: 10.3389/fpls.2019.00930. eCollection 2019. Front Plant Sci. 2019. PMID: 31379907 Free PMC article.
-
Identification and functional characterization of bidirectional gene pairs and their intergenic regions in cotton.BMC Plant Biol. 2024 Sep 4;24(1):829. doi: 10.1186/s12870-024-05548-w. BMC Plant Biol. 2024. PMID: 39232709 Free PMC article.
-
Suitability of Real-Time PCR Methods for New Genomic Technique Detection in the Context of the European Regulations: A Case Study in Arabidopsis.Int J Mol Sci. 2025 Apr 2;26(7):3308. doi: 10.3390/ijms26073308. Int J Mol Sci. 2025. PMID: 40244157 Free PMC article.
-
Unraveling the regulatory dynamics of bidirectional promoters for modulating gene co-expression and metabolic flux in Saccharomyces cerevisiae.Nucleic Acids Res. 2025 Jun 6;53(11):gkaf511. doi: 10.1093/nar/gkaf511. Nucleic Acids Res. 2025. PMID: 40503683 Free PMC article.
-
Specificity Testing for NGT PCR-Based Detection Methods in the Context of the EU GMO Regulations.Foods. 2023 Nov 28;12(23):4298. doi: 10.3390/foods12234298. Foods. 2023. PMID: 38231759 Free PMC article.
References
-
- James C. Global Status of Commercialized Biotech/GM Crops, 2017. Volume 36. International Service for the Acquisition of Agri-Biotech Applications; New York, NY, USA: 2018. pp. 1–8.
-
- Kumari M., Rai A.K., Devanna B.N., Singh P.K., Kapoor R., Rajashekara H., Prakash G., Sharma V., Sharma T.R. Co-transformation mediated stacking of blast resistance genes Pi54 and Pi54rh in rice provides broad spectrum resistance against Magnaporthe oryzae. Plant Cell Rep. 2017;36:1747–1755. doi: 10.1007/s00299-017-2189-x. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

