Unraveling the effect of silent, intronic and missense mutations on VWF splicing: contribution of next generation sequencing in the study of mRNA
- PMID: 30361419
- PMCID: PMC6395343
- DOI: 10.3324/haematol.2018.203166
Unraveling the effect of silent, intronic and missense mutations on VWF splicing: contribution of next generation sequencing in the study of mRNA
Abstract
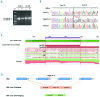
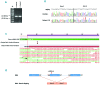
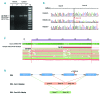
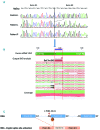
Large studies in von Willebrand disease patients, including Spanish and Portuguese registries, led to the identification of >250 different mutations. It is a challenge to determine the pathogenic effect of potential splice site mutations on VWF mRNA. This study aimed to elucidate the true effects of 18 mutations on VWF mRNA processing, investigate the contribution of next-generation sequencing to in vivo mRNA study in von Willebrand disease, and compare the findings with in silico prediction. RNA extracted from patient platelets and leukocytes was amplified by RT-PCR and sequenced using Sanger and next generation sequencing techniques. Eight mutations affected VWF splicing: c.1533+1G>A, c.5664+2T>C and c.546G>A (p.=) prompted exon skipping; c.3223-7_3236dup and c.7082-2A>G resulted in activation of cryptic sites; c.3379+1G>A and c.7437G>A) demonstrated both molecular pathogenic mechanisms simultaneously; and the p.Cys370Tyr missense mutation generated two aberrant transcripts. Of note, the complete effect of three mutations was provided by next generation sequencing alone because of low expression of the aberrant transcripts. In the remaining 10 mutations, no effect was elucidated in the experiments. However, the differential findings obtained in platelets and leukocytes provided substantial evidence that four of these would have an effect on VWF levels. In this first report using next generation sequencing technology to unravel the effects of VWF mutations on splicing, the technique yielded valuable information. Our data bring to light the importance of studying the effect of synonymous and missense mutations on VWF splicing to improve the current knowledge of the molecular mechanisms behind von Willebrand disease. clinicaltrials.gov identifier:02869074.
Copyright© 2019 Ferrata Storti Foundation.
Figures

Similar articles
-
The study of the effect of splicing mutations in von Willebrand factor using RNA isolated from patients' platelets and leukocytes.J Thromb Haemost. 2011 Apr;9(4):679-88. doi: 10.1111/j.1538-7836.2011.04204.x. J Thromb Haemost. 2011. PMID: 21251206
-
A synonymous (c.3390C>T) or a splice-site (c.3380-2A>G) mutation causes exon 26 skipping in four patients with von Willebrand disease (2A/IIE).J Thromb Haemost. 2013 Jul;11(7):1251-9. doi: 10.1111/jth.12280. J Thromb Haemost. 2013. PMID: 23621778
-
Unraveling the molecular basis underlying nine putative splice site variants of von Willebrand factor.Hum Mutat. 2022 Feb;43(2):215-227. doi: 10.1002/humu.24312. Epub 2021 Dec 17. Hum Mutat. 2022. PMID: 34882887
-
Laboratory diagnosis of von Willebrand disease type 1/2E (2A subtype IIE), type 1 Vicenza and mild type 1 caused by mutations in the D3, D4, B1-B3 and C1-C2 domains of the von Willebrand factor gene. Role of von Willebrand factor multimers and the von Willebrand factor propeptide/antigen ratio.Acta Haematol. 2009;121(2-3):128-38. doi: 10.1159/000214853. Epub 2009 Jun 8. Acta Haematol. 2009. PMID: 19506359 Review.
-
Molecular genetics of type 2 von Willebrand disease.Int J Hematol. 2002 Jan;75(1):9-18. doi: 10.1007/BF02981973. Int J Hematol. 2002. PMID: 11843298 Review.
Cited by
-
Molecular basis of inherited protein C deficiency results from genetic variations in the signal peptide and propeptide regions.J Thromb Haemost. 2023 Nov;21(11):3124-3137. doi: 10.1016/j.jtha.2023.06.021. Epub 2023 Jun 29. J Thromb Haemost. 2023. PMID: 37393002 Free PMC article.
-
Cryptic non-canonical splice site activation is part of the mechanism that abolishes multimer organization in the c.2269_2270del von Willebrand factor.Haematologica. 2020 Apr;105(4):1120-1128. doi: 10.3324/haematol.2019.222679. Epub 2019 Jul 18. Haematologica. 2020. PMID: 31320553 Free PMC article.
-
Novel De Novo Intronic Variant of SYNGAP1 Associated With the Neurodevelopmental Disorders.Mol Genet Genomic Med. 2025 Feb;13(2):e70066. doi: 10.1002/mgg3.70066. Mol Genet Genomic Med. 2025. PMID: 39878419 Free PMC article.
-
The CDH1 c.1901C>T Variant: A Founder Variant in the Portuguese Population with Severe Impact in mRNA Splicing.Cancers (Basel). 2021 Sep 4;13(17):4464. doi: 10.3390/cancers13174464. Cancers (Basel). 2021. PMID: 34503274 Free PMC article.
-
POLR3A rare variants in a patient with intellectual disability, ataxic gait and cortical malformations: a case-report.Ital J Pediatr. 2025 Jun 15;51(1):191. doi: 10.1186/s13052-025-02024-5. Ital J Pediatr. 2025. PMID: 40518520 Free PMC article.
References
-
- Rodeghiero F, Castaman G, Dini E. Epidemiological investigation of the prevalence of von Willebrand’s disease. Blood. 1987;69(2):454–459. - PubMed
-
- Batlle J, Perez-Rodriguez A, Corrales I, et al. Molecular and clinical profile of von Willebrand disease in Spain (PCM-EVW-ES): Proposal for a new diagnostic paradigm. Thromb Haemost. 2016;115(1):40–50. - PubMed
-
- Fidalgo T, Salvado R, Corrales I, et al. Genotype-phenotype correlation in a cohort of Portuguese patients comprising the entire spectrum of VWD types: impact of NGS. Thromb Haemost. 2016;116(1):17–31. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

