Expansion and differentiation of human hepatocyte-derived liver progenitor-like cells and their use for the study of hepatotropic pathogens
- PMID: 30361550
- PMCID: PMC6318298
- DOI: 10.1038/s41422-018-0103-x
Expansion and differentiation of human hepatocyte-derived liver progenitor-like cells and their use for the study of hepatotropic pathogens
Abstract
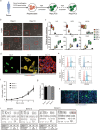
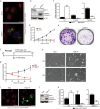
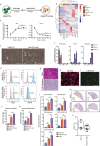
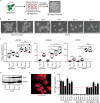
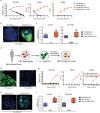
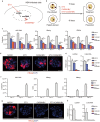
The study of pathophysiological mechanisms in human liver disease has been constrained by the inability to expand primary hepatocytes in vitro while maintaining proliferative capacity and metabolic function. We and others have previously shown that mouse mature hepatocytes can be converted to liver progenitor-like cells in vitro with defined chemical factors. Here we describe a protocol achieving efficient conversion of human primary hepatocytes into liver progenitor-like cells (HepLPCs) through delivery of developmentally relevant cues, including NAD + -dependent deacetylase SIRT1 signaling. These HepLPCs could be expanded significantly during in vitro passage. The expanded cells can readily be converted back into metabolically functional hepatocytes in vitro and upon transplantation in vivo. Under three-dimensional culture conditions, differentiated cells generated from HepLPCs regained the ability to support infection or reactivation of hepatitis B virus (HBV). Our work demonstrates the utility of the conversion between hepatocyte and liver progenitor-like cells for studying HBV biology and antiviral therapies. These findings will facilitate the study of liver diseases and regenerative medicine.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Junge N., Sharma A. D. & Ott M. About cytokeratin 19 and the drivers of liver regeneration. J. Hepatol68, 1 (2017). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

