The Hierarchical Modular Structure of HER2+ Breast Cancer Network
- PMID: 30364267
- PMCID: PMC6193406
- DOI: 10.3389/fphys.2018.01423
The Hierarchical Modular Structure of HER2+ Breast Cancer Network
Abstract
HER2-enriched breast cancer is a complex disease characterized by the overexpression of the ERBB2 amplicon. While the effects of this genomic aberration on the pathology have been studied, genome-wide deregulation patterns in this subtype of cancer are also observed. A novel approach to the study of this malignant neoplasy is the use of transcriptional networks. These networks generally exhibit modular structures, which in turn may be associated to biological processes. This modular regulation of biological functions may also exhibit a hierarchical structure, with deeper levels of modular organization accounting for more specific functional regulation. In this work, we identified the most probable (maximum likelihood) model of the hierarchical modular structure of the HER2-enriched transcriptional network as reconstructed from gene expression data, and analyzed the statistical associations of modules and submodules to biological functions. We found modular structures, independent from direct ERBB2 amplicon regulation, involved in different biological functions such as signaling, immunity, and cellular morphology. Higher resolution submodules were identified in more specific functions, such as micro-RNA regulation and the activation of viral-like immune response. We propose the approach presented here as one that may help to unveil mechanisms involved in the development of the pathology.
Keywords: breast cancer; gene regulatory networks; genetic; modular networks; molecular subtypes; signaling pathways; transcription.
Figures

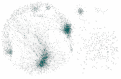




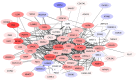


References
-
- Amini A. A., Chen A., Bickel P. J., Levina E. (2013). Pseudo-likelihood methods for community detection in large sparse networks. Ann. Statist. 41, 2097–2122. 10.1214/13-AOS1138 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

