Molecular Cloning and Functional Characterization of the Dehydrin (IpDHN) Gene From Ipomoea pes-caprae
- PMID: 30364314
- PMCID: PMC6193111
- DOI: 10.3389/fpls.2018.01454
Molecular Cloning and Functional Characterization of the Dehydrin (IpDHN) Gene From Ipomoea pes-caprae
Abstract
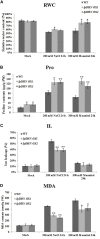
Dehydrin (DHN) genes can be rapidly induced to offset water deficit stresses in plants. Here, we reported on a dehydrin gene (IpDHN) related to salt tolerance isolated from Ipomoea pes-caprae L. (Convolvulaceae). The IpDHN protein shares a relatively high homology with Arabidopsis dehydrin ERD14 (At1g76180). IpDHN was shown to have a cytoplasmic localization pattern. Quantitative RT-PCR analyses indicated that IpDHN was differentially expressed in most organs of I. pes-caprae plants, and its expression level increased after salt, osmotic stress, oxidative stress, cold stress and ABA treatments. Analysis of the 974-bp promoter of IpDHN identified distinct cis-acting regulatory elements, including an MYB binding site (MBS), ABRE (ABA responding)-elements, Skn-1 motif, and TC-rich repeats. The induced expression of IpDHN in Escherichia coli indicated that IpDHN might be involved in salt, drought, osmotic, and oxidative stresses. We also generated transgenic Arabidopsis lines that over-expressed IpDHN. The transgenic Arabidopsis plants showed a significant enhancement in tolerance to salt/drought stresses, as well as less accumulation of hydrogen peroxide (H2O2) and the superoxide radical (O2 -), accompanied by increasing activity of the antioxidant enzyme system in vivo. Under osmotic stresses, the overexpression of IpDHN in Arabidopsis can elevate the expression of ROS-related and stress-responsive genes and can improve the ROS-scavenging ability. Our results indicated that IpDHN is involved in cellular responses to salt and drought through a series of pleiotropic effects that are likely involved in ROS scavenging and therefore influence the physiological processes of microorganisms and plants exposed to many abiotic stresses.
Keywords: Ipomoea pes-caprae L.; dehydrin; drought; promoter; salt.
Figures

 ” shows the position of IpDHN in the phylogenetic tree.
” shows the position of IpDHN in the phylogenetic tree.









References
-
- Alsheikh M. K., Svensson J. T., Randall S. K. (2005). Phosphorylation regulated ion-binding is a property shared by the acidic subclass dehydrins. Plant Cell Environ. 28 1114–1122. 10.1111/j.1365-3040.2005.01348.x - DOI
LinkOut - more resources
Full Text Sources

