Effect Sizes of Somatic Mutations in Cancer
- PMID: 30365005
- PMCID: PMC6235682
- DOI: 10.1093/jnci/djy168
Effect Sizes of Somatic Mutations in Cancer
Abstract
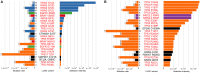
A major goal of cancer biology is determination of the relative importance of the genetic alterations that confer selective advantage to cancer cells. Tumor sequence surveys have frequently ranked the importance of substitutions to cancer growth by P value or a false-discovery conversion thereof. However, P values are thresholds for belief, not metrics of effect. Their frequent misuse as metrics of effect has often been vociferously decried, even in cases when the only attributable mistake was omission of effect sizes. Here, we propose an appropriate ranking-the cancer effect size, which is the selection intensity for somatic variants in cancer cell lineages. The selection intensity is a metric of the survival and reproductive advantage conferred by mutations in somatic tissue. Thus, they are of fundamental importance to oncology, and have immediate relevance to ongoing decision making in precision medicine tumor boards, to the selection and design of clinical trials, to the targeted development of pharmaceuticals, and to basic research prioritization. Within this commentary, we first discuss the scope of current methods that rank confidence in the overrepresentation of specific mutated genes in cancer genomes. Then we bring to bear recent advances that draw upon an understanding of the development of cancer as an evolutionary process to estimate the effect size of somatic variants leading to cancer. We demonstrate the estimation of the effect sizes of all recurrent single nucleotide variants in 22 cancer types, quantifying relative importance within and between driver genes.
Figures


References
-
- Martincorena I, Raine KM, Gerstung M, et al. Universal patterns of selection in cancer and somatic tissues. Cell. 2017;171(5):1029–1041. doi:10.1016/j.cell.2017.09.042 - DOI - PMC - PubMed
-
- Armenia J, Wankowicz SAM, Liu D, et al. The long tail of oncogenic drivers in prostate cancer. Nat Genet. 2018;50(5):645–651. doi:10.1038/s41588-018-0078-z - DOI - PMC - PubMed
-
- Kan Z, Jaiswal BS, Stinson J, et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466(7308):869–873. - PubMed
-
- Zang ZJ, Cutcutache I, Poon SL, et al. Exome sequencing of gastric adenocarcinoma identifies recurrent somatic mutations in cell adhesion and chromatin remodeling genes. Nat Genet. 2012;44(5):570–574. - PubMed

