CXXC1 is not essential for normal DNA double-strand break formation and meiotic recombination in mouse
- PMID: 30365547
- PMCID: PMC6221362
- DOI: 10.1371/journal.pgen.1007657
CXXC1 is not essential for normal DNA double-strand break formation and meiotic recombination in mouse
Abstract
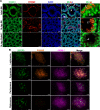
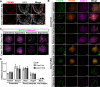
In most mammals, including mice and humans, meiotic recombination is determined by the meiosis specific histone methytransferase PRDM9, which binds to specific DNA sequences and trimethylates histone 3 at lysine-4 and lysine-36 at the adjacent nucleosomes. These actions ensure successful DNA double strand break formation and repair that occur on the proteinaceous structure forming the chromosome axis. The process of hotspot association with the axis after their activation by PRDM9 is poorly understood. Previously, we and others have identified CXXC1, an ortholog of S. cerevisiae Spp1 in mammals, as a PRDM9 interactor. In yeast, Spp1 is a histone methyl reader that links H3K4me3 sites with the recombination machinery, promoting DSB formation. Here, we investigated whether CXXC1 has a similar function in mouse meiosis. We created two Cxxc1 conditional knockout mouse models to deplete CXXC1 generally in germ cells, and before the onset of meiosis. Surprisingly, male knockout mice were fertile, and the loss of CXXC1 in spermatocytes had no effect on PRDM9 hotspot trimethylation, double strand break formation or repair. Our results demonstrate that CXXC1 is not an essential link between PRDM9-activated recombination hotspot sites and DSB machinery and that the hotspot recognition pathway in mouse is independent of CXXC1.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

