Integrin but not CEACAM receptors are dispensable for Helicobacter pylori CagA translocation
- PMID: 30365569
- PMCID: PMC6231679
- DOI: 10.1371/journal.ppat.1007359
Integrin but not CEACAM receptors are dispensable for Helicobacter pylori CagA translocation
Abstract
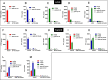
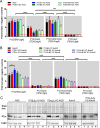
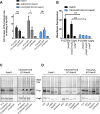
Translocation of the Helicobacter pylori (Hp) cytotoxin-associated gene A (CagA) effector protein via the cag-Type IV Secretion System (cag-T4SS) into host cells is a hallmark of infection with Hp and a major risk factor for severe gastric diseases, including gastric cancer. To mediate the injection of CagA, Hp uses a membrane-embedded syringe-like molecular apparatus extended by an external pilus-like rod structure that binds host cell surface integrin heterodimers. It is still largely unclear how the interaction of the cag-T4SS finally mediates translocation of the CagA protein into the cell cytoplasm. Recently certain carcinoembryonic antigen-related cell adhesion molecules (CEACAMs), acting as receptor for the Hp outer membrane adhesin HopQ, have been identified to be involved in the process of CagA host cell injection. Here, we applied the CRISPR/Cas9-knockout technology to generate defined human gastric AGS and KatoIII integrin knockout cell lines. Although confocal laser scanning microscopy revealed a co-localization of Hp and β1 integrin heterodimers on gastric epithelial cells, Hp infection studies using the quantitative and highly sensitive Hp β-lactamase reporter system clearly show that neither β1 integrin heterodimers (α1β1, α2β1 or α5β1), nor any other αβ integrin heterodimers on the cell surface are essential for CagA translocation. In contrast, deletion of the HopQ adhesin in Hp, or the simultaneous knockout of the receptors CEACAM1, CEACAM5 and CEACAM6 in KatoIII cells abolished CagA injection nearly completely, although bacterial binding was only reduced to 50%. These data provide genetic evidence that the cag-T4SS-mediated interaction of Hp with cell surface integrins on human gastric epithelial cells is not essential for CagA translocation, but interaction of Hp with CEACAM receptors is facilitating CagA translocation by the cag-T4SS of this important microbe.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Comment in
-
Different roles of integrin-β1 and integrin-αv for type IV secretion of CagA versus cell elongation phenotype and cell lifting by Helicobacter pylori.PLoS Pathog. 2020 Jul 21;16(7):e1008135. doi: 10.1371/journal.ppat.1008135. eCollection 2020 Jul. PLoS Pathog. 2020. PMID: 32692784 Free PMC article. No abstract available.
References
-
- Odenbreit S, Püls J, Sedlmaier B, Gerland E, Fischer W, Haas R. Translocation of Helicobacter pylori CagA into gastric epithelial cells by type IV secretion. Science. 2000;287:1497–500. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

