Normalized Quantitative PCR Measurements as Predictors for Ethene Formation at Sites Impacted with Chlorinated Ethenes
- PMID: 30365883
- PMCID: PMC6945293
- DOI: 10.1021/acs.est.8b04373
Normalized Quantitative PCR Measurements as Predictors for Ethene Formation at Sites Impacted with Chlorinated Ethenes
Abstract
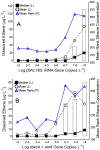
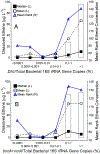
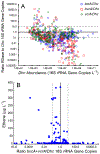
Quantitative PCR (qPCR) targeting Dehalococcoides mccartyi ( Dhc) biomarker genes supports effective management at sites impacted with chlorinated ethenes. To establish correlations between Dhc biomarker gene abundances and ethene formation (i.e., detoxification), 859 groundwater samples representing 62 sites undergoing monitored natural attenuation or enhanced remediation were analyzed. Dhc 16S rRNA genes and the vinyl chloride (VC) reductive dehalogenase genes bvcA and vcrA were detected in 88% and 61% of samples, respectively, from wells with ethene. Dhc 16S rRNA, bvcA, vcrA, and tceA (implicated in cometabolic reductive VC dechlorination) gene abundances all positively correlated with ethene formation. Significantly greater ethene concentrations were observed when Dhc 16S rRNA gene and VC RDase gene abundances exceeded 107 and 106 copies L-1, respectively, and when Dhc 16S rRNA- and bvcA + vcrA-to-total bacterial 16S rRNA gene ratios exceeded 0.1%. Dhc 16S rRNA gene-to- vcrA/ bvcA ratios near unity also indicated elevated ethene; however, no increased ethene was observed in 19 wells where vcrA and/or bvcA gene copy numbers exceeded Dhc cell numbers 10- to 10 000-fold. Approximately one-third of samples with detectable ethene lacked bvcA, vcrA, and tceA, suggesting that comprehensive understanding of VC detoxification biomarkers has not been achieved. Although the current biomarker suite is incomplete, the data analysis corroborates the value of the available Dhc DNA biomarkers for prognostic and diagnostic groundwater monitoring at sites impacted with chlorinated ethenes.
Figures

References
-
- Löffler FE; Ritalahti KM; Zinder SH Dehalococcoides and reductive dechlorination of chlorinated solvents In SERDP ESTCP Environmental Remediation Technology; Stroo HF, Leeson A, Ward CH, Eds.; Springer: New York, NY, 2013; Vol. Bioaugmentation for Groundwater Remediation; pp 39–88.
-
- Maymó-Gatell X; Chien Y; Gossett JM; Zinder SH Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene. Science 1997, 276 (5318), 1568–1571. - PubMed
-
- He J; Ritalahti KM; Yang K-L; Koenigsberg SS; Löffler FE Detoxification of vinyl chloride to ethene coupled to growth of an anaerobic bacterium. Nature 2003, 424, 62–65. - PubMed
-
- Ritalahti KM; Hatt JK; Lugmayr V; Henn K; Petrovskis EA; Ogles DM; Davis GA; Yeager CM; Lebrón CA; Löffler FE Comparing on-site to off-site biomass collection for Dehalococcoides biomarker gene quantification to predict in situ chlorinated ethene detoxification potential. Environ. Sci. Technol 2010, 44 (13), 5127–5133. - PubMed
-
- Löffler FE; Yan J; Ritalahti KM; Adrian L; Edwards EA; Konstantinidis KT; Müller JA; Fullerton H; Zinder S; Spormann AM Dehalococcoides mccartyi gen. nov., sp. nov., obligate organohalide-respiring anaerobic bacteria relevant to halogen cycling and bioremediation, belong to a novel bacterial class, Dehalococcoidia classis nov., order Dehalococcoidales ord. nov. and family Dehalococcoi-daceae fam. nov., within the phylum Chlorof lexi. Int. J. Syst. Evol. Microbiol 2013, 63 (2), 625–635. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

