Highly sensitive detection of DNA hypermethylation in melanoma cancer cells
- PMID: 30366258
- PMCID: PMC6245664
- DOI: 10.1016/j.bios.2018.10.018
Highly sensitive detection of DNA hypermethylation in melanoma cancer cells
Abstract
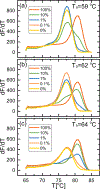
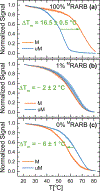
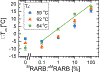
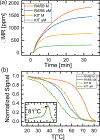
Aberrant hypermethylation of CpG islands in the promoter region of tumor suppressor genes is a promising biomarker for early cancer detection. This methylation status is reflected in the methylation pattern of ctDNA shed from the primary tumor; however, to realize the full clinical utility of ctDNA methylation detection via liquid biopsy for early cancer diagnosis, improvements in the sensitivity and multiplexability of existing technologies must be improved. Additionally, the assay must be cheap and easy to perform in a clinical setting. We report the integration of methylation specific PCR (MSP) to melt curve analysis on giant magnetoresistive (GMR) biosensors to greatly enhance the sensitivity of our DNA hybridization assay for methylation detection. Our GMR sensor is functionalized with synthetic DNA probes that target methylated or unmethylated CpG sites in the MSP amplicon, and measures the difference in melting temperature (Tm) between the two probes (ΔTm), giving an analytical limit of detection down to 0.1% methylated DNA in solution. Additionally, linear regression of ΔTm's for serial dilutions of methylated:unmethylated mixtures allows for quantification of methylation percentage, which could have diagnostic and prognostic utility. Lastly, we performed multiplexed MSP on two different genes, and show the ability of our GMR assay to resolve this mixture, despite their amplicons' overlapping Tm's in standard EvaGreen melt analysis. The multiplexing ability of our assay and its enhanced sensitivity, without necessitating deep sequencing, represent important steps toward realizing an assay for the detection of methylated ctDNA in plasma for early cancer detection in a clinical setting.
Keywords: Array; Cancer; DNA melting; GMR sensors; Methylation specific PCR; PCR.
Copyright © 2018 Elsevier B.V. All rights reserved.
Conflict of interest statement
Conflict of Interest
The authors declare the following competing financial interest(s): S.X.W. have related patents or patent applications assigned to Stanford University and out-licensed for potential commercialization. S.X.W. has stock or stock options in MagArray, Inc., which has licensed relevant patents from Stanford University for commercialization of GMR nanosensor chips.
Figures

References
-
- Aravanis AM, Lee M, Klausner RD, 2017. Next-Generation Sequencing of Circulating Tumor DNA for Early Cancer Detection. Cell 168, 571–574. https://doi.org/10.10167j.cell.2017.01.030 - PubMed
-
- Baylin SB, 2005. DNA methylation and gene silencing in cancer. Nat. Rev. Clin. Oncol. 2, S4–S11. https://doi.org/10.1038/ncponc0354 - DOI - PubMed
-
- Belinsky SA, 2005. Silencing of genes by promoter hypermethylation: key event in rodent and human lung cancer. Carcinogenesis 26, 1481–1487. https://doi.org/10.1093/carcin/bgi020 - DOI - PubMed
-
- Calapre L, Warburton L, Millward M, Ziman M, Gray ES, 2017. Circulating tumour DNA (ctDNA) as a liquid biopsy for melanoma. Cancer Lett. 404, 62–69. https://doi.org/10.1016/j.canlet.2017.06.030 - DOI - PubMed
-
- Chimonidou M, Strati A, Malamos N, Kouneli S, Georgoulias V, Lianidou E, Chimonidou M, Strati A, Malamos N, Kouneli S, Georgoulias V, Lianidou E, 2017. Direct comparison study of DNA methylation markers in EpCAM-positive circulating tumour cells, corresponding circulating tumour DNA, and paired primary tumours in breast cancer. Oncotarget 8, 72054–72068. https://doi.org/10.18632/oncotarget.18679 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

