Expression of human miR-200b-3p and -200c-3p in cytomegalovirus-infected tissues
- PMID: 30366960
- PMCID: PMC6435554
- DOI: 10.1042/BSR20180961
Expression of human miR-200b-3p and -200c-3p in cytomegalovirus-infected tissues
Abstract
Human cytomegalovirus (HCMV) infection can cause inflammatory tissue-invasive end-organ diseases upon lytic replication. In humans, mature miR-200b-3p and -200c-3p suppress the synthesis of HCMV immediate early 2 (IE2) protein by binding to the 3'-UTR of the mRNA encoded by the unique long (UL) 122-123 region in human foreskin fibroblasts and pre-transplant peripheral blood mononuclear cells stimulated with HCMV. The present study aimed to quantitate the expression of Homo sapiens (hsa)-miR-200b-3p and 200c-3p in HCMV-infected tissues. We collected 240 HCMV-infected and 154 HCMV-non-infected, formalin-fixed, paraffin-embedded tissue samples of the gastrointestinal (GI) tract and bronchi/lungs. MiRNAs, HCMV, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were quantitated by quantitative reverse transcription-PCR (qRT-PCR) and quantitative PCR (qPCR) on the basis of standard curves generated using miRNA mimics, the HCMV strain from National Institute for Biological Standards and Control (NIBSC) 09/162, and GAPDH control. To avoid the effect of cell counts on the qRT-PCR and qPCR results, the data were normalized to GAPDH levels. HCMV-infected tissues had significantly lower levels of 200b-3p/GAPDH (3.03 ± 1.50 compared with 3.98 ± 1.08 log10 copies/μl, P<0.001) and 200c-3p/GAPDH (4.67 ± 1.84 compared with 6.35 ± 1.47 log10 copies/μl, P<0.001) than normal tissues. The values for 200b-3p/GAPDH (r = -0.51, P<0.001) and 200c-3p/GAPDH (r = -0.54, P<0.001) were significantly inversely correlated with HCMV load. Low tissue levels of 200b-3p and 200c-3p in humans are associated with cytopathic inflammation due to HCMV infection.
Keywords: cytomegalovirus; immediate early protein 2; microRNA; tissues.
© 2018 The Author(s).
Conflict of interest statement
The authors declare that there are no competing interests associated with the manuscript.
Figures


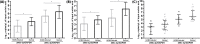

Similar articles
-
Human MicroRNA Responses Predict Cytomegalovirus Replication Following Solid Organ Transplantation.J Infect Dis. 2017 Feb 15;215(4):537-546. doi: 10.1093/infdis/jiw596. J Infect Dis. 2017. PMID: 28003351
-
Human MicroRNAs Attenuate the Expression of Immediate Early Proteins and HCMV Replication during Lytic and Latent Infection in Connection with Enhancement of Phosphorylated RelA/p65 (Serine 536) That Binds to MIEP.Int J Mol Sci. 2022 Mar 2;23(5):2769. doi: 10.3390/ijms23052769. Int J Mol Sci. 2022. PMID: 35269913 Free PMC article.
-
Human cytomegalovirus latent infection alters the expression of cellular and viral microRNA.Gene. 2014 Feb 25;536(2):272-8. doi: 10.1016/j.gene.2013.12.012. Epub 2013 Dec 18. Gene. 2014. PMID: 24361963
-
Roles of host and viral microRNAs in human cytomegalovirus biology.Virus Res. 2011 May;157(2):180-92. doi: 10.1016/j.virusres.2010.10.011. Epub 2010 Oct 20. Virus Res. 2011. PMID: 20969901 Free PMC article. Review.
-
Human cytomegalovirus (HCMV)-encoded microRNAs: potential biomarkers and clinical applications.RNA Biol. 2021 Dec;18(12):2194-2202. doi: 10.1080/15476286.2021.1930757. Epub 2021 May 26. RNA Biol. 2021. PMID: 34039247 Free PMC article. Review.
Cited by
-
Baicalin attenuates LPS-induced alveolar type II epithelial cell A549 injury by attenuation of the FSTL1 signaling pathway via increasing miR-200b-3p expression.Innate Immun. 2021 May;27(4):294-312. doi: 10.1177/17534259211013887. Epub 2021 May 18. Innate Immun. 2021. PMID: 34000873 Free PMC article.
-
The mechanism of the Nfe2l2/Hmox1 signaling pathway in ferroptosis regulation in acute compartment syndrome.J Biochem Mol Toxicol. 2023 Jan;37(1):e23228. doi: 10.1002/jbt.23228. Epub 2022 Oct 4. J Biochem Mol Toxicol. 2023. PMID: 36193742 Free PMC article.
-
Research progress on miRNAs function in the interaction between human infectious viruses and hosts: A review.Biomol Biomed. 2024 Oct 17;24(6):1452-1462. doi: 10.17305/bb.2024.10821. Biomol Biomed. 2024. PMID: 39101759 Free PMC article. Review.
-
Human Cytomegalovirus Induced Aberrant Expression of Non-coding RNAs.Front Microbiol. 2022 Jun 13;13:918213. doi: 10.3389/fmicb.2022.918213. eCollection 2022. Front Microbiol. 2022. PMID: 35770158 Free PMC article. Review.
-
Characterization of microRNA Expression Profiles of Murine Female Genital Tracts Following Nippostrongylus brasiliensis and Herpes Simplex Virus Type 2 Co-Infection.Microorganisms. 2025 Jul 24;13(8):1734. doi: 10.3390/microorganisms13081734. Microorganisms. 2025. PMID: 40871238 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

