Genomic and Transcriptomic Basis of Hanseniaspora vineae's Impact on Flavor Diversity and Wine Quality
- PMID: 30366992
- PMCID: PMC6293095
- DOI: 10.1128/AEM.01959-18
Genomic and Transcriptomic Basis of Hanseniaspora vineae's Impact on Flavor Diversity and Wine Quality
Abstract
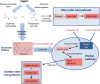
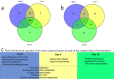
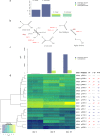
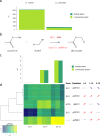
Hanseniaspora is the main genus of the apiculate yeast group that represents approximately 70% of the grape-associated microflora. Hanseniaspora vineae is emerging as a promising species for quality wine production compared to other non-Saccharomyces species. Wines produced by H. vineae with Saccharomyces cerevisiae consistently exhibit more intense fruity flavors and complexity than wines produced by S. cerevisiae alone. In this work, genome sequencing, assembling, and phylogenetic analysis of two strains of H. vineae showed that it is a member of the Saccharomyces complex and it diverged before the whole-genome duplication (WGD) event from this clade. Specific flavor gene duplications and absences were identified in the H. vineae genome compared to 14 fully sequenced industrial S. cerevisiae genomes. The increased formation of 2-phenylethyl acetate and phenylpropanoids such as 2-phenylethyl and benzyl alcohols might be explained by gene duplications of H. vineae aromatic amino acid aminotransferases (ARO8 and ARO9) and phenylpyruvate decarboxylases (ARO10). Transcriptome and aroma profiles under fermentation conditions confirmed these genes were highly expressed at the beginning of stationary phase coupled to the production of their related compounds. The extremely high level of acetate esters produced by H. vineae compared to that by S. cerevisiae is consistent with the identification of six novel proteins with alcohol acetyltransferase (AATase) domains. The absence of the branched-chain amino acid transaminases (BAT2) and acyl coenzyme A (acyl-CoA)/ethanol O-acyltransferases (EEB1) genes correlates with H. vineae's reduced production of branched-chain higher alcohols, fatty acids, and ethyl esters, respectively. Our study provides sustenance for understanding and potentially utilizing genes that determine fermentation aromas.IMPORTANCE The huge diversity of non-Saccharomyces yeasts in grapes is dominated by the apiculate genus Hanseniaspora Two native strains of Hanseniaspora vineae applied to winemaking because of their high oenological potential in aroma and fermentation performance were selected to obtain high-quality genomes. Here, we present a phylogenetic analysis and the complete transcriptome and aroma metabolome of H. vineae during three fermentation steps. This species produced significantly richer flavor compound diversity than Saccharomyces, including benzenoids, phenylpropanoids, and acetate-derived compounds. The identification of six proteins, different from S. cerevisiae ATF, with diverse acetyltransferase domains in H. vineae offers a relevant source of native genetic variants for this enzymatic activity. The discovery of benzenoid synthesis capacity in H. vineae provides a new eukaryotic model to dilucidate an alternative pathway to that catalyzed by plants' phenylalanine lyases.
Keywords: Illumina; flavor compounds; genome; metabolome; transcriptome; wine aroma.
Copyright © 2018 American Society for Microbiology.
Figures

References
-
- Martin V, Valera MJ, Medina K, Boido E, Carrau F. 2018. Oenological impact of the Hanseniaspora/Kloeckera yeast genus on wines–a review. Fermentation 4:76. doi:10.3390/fermentation4030076. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

