Common helical V1V2 conformations of HIV-1 Envelope expose the α4β7 binding site on intact virions
- PMID: 30367034
- PMCID: PMC6203816
- DOI: 10.1038/s41467-018-06794-x
Common helical V1V2 conformations of HIV-1 Envelope expose the α4β7 binding site on intact virions
Abstract
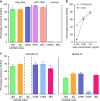
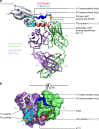
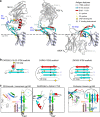
The α4β7 integrin is a non-essential HIV-1 adhesion receptor, bound by the gp120 V1V2 domain, facilitating rapid viral dissemination into gut-associated lymphoid tissues. Antibodies blocking this interaction early in infection can improve disease outcome, and V1V2-targeted antibodies were correlated with moderate efficacy reported from the RV144 HIV-1 vaccine trial. Monoclonal α4β7-blocking antibodies recognise two slightly different helical V2 conformations, and current structural data suggests their binding sites are occluded in prefusion envelope trimers. Here, we report cocrystal structures of two α4β7-blocking antibodies from an infected donor complexed with scaffolded V1V2 or V2 peptides. Both antibodies recognised the same helix-coil V2 conformation as RV144 antibody CH58, identifying a frequently sampled alternative conformation of full-length V1V2. In the context of Envelope, this α-helical form of V1V2 displays highly exposed α4β7-binding sites, potentially providing a functional role for non-native Envelope on virion or infected cell surfaces in HIV-1 dissemination, pathogenesis, and vaccine design.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Identification of New Regions in HIV-1 gp120 Variable 2 and 3 Loops that Bind to α4β7 Integrin Receptor.PLoS One. 2015 Dec 1;10(12):e0143895. doi: 10.1371/journal.pone.0143895. eCollection 2015. PLoS One. 2015. PMID: 26625359 Free PMC article. Clinical Trial.
-
Select gp120 V2 domain specific antibodies derived from HIV and SIV infection and vaccination inhibit gp120 binding to α4β7.PLoS Pathog. 2018 Aug 28;14(8):e1007278. doi: 10.1371/journal.ppat.1007278. eCollection 2018 Aug. PLoS Pathog. 2018. PMID: 30153309 Free PMC article.
-
Plasticity and Epitope Exposure of the HIV-1 Envelope Trimer.J Virol. 2017 Aug 10;91(17):e00410-17. doi: 10.1128/JVI.00410-17. Print 2017 Sep 1. J Virol. 2017. PMID: 28615206 Free PMC article.
-
The HIV-1 gp120 V1V2 loop: structure, function and importance for vaccine development.Expert Rev Vaccines. 2014 Dec;13(12):1489-500. doi: 10.1586/14760584.2014.951335. Epub 2014 Aug 28. Expert Rev Vaccines. 2014. PMID: 25163695 Review.
-
Integrin α4β7 in HIV-1 infection: A critical review.J Leukoc Biol. 2020 Aug;108(2):627-632. doi: 10.1002/JLB.4MR0120-208R. Epub 2020 Apr 9. J Leukoc Biol. 2020. PMID: 32272507 Review.
Cited by
-
Vaccine-induced V1V2-specific antibodies control and or protect against infection with HIV, SIV and SHIV.Curr Opin HIV AIDS. 2019 Jul;14(4):309-317. doi: 10.1097/COH.0000000000000551. Curr Opin HIV AIDS. 2019. PMID: 30994501 Free PMC article. Review.
-
AAV-Mediated Expression of Broadly Neutralizing and Vaccine-like Antibodies Targeting the HIV-1 Envelope V2 Region.Mol Ther Methods Clin Dev. 2019 Jun 12;14:100-112. doi: 10.1016/j.omtm.2019.06.002. eCollection 2019 Sep 13. Mol Ther Methods Clin Dev. 2019. PMID: 31334303 Free PMC article.
-
To bnAb or Not to bnAb: Defining Broadly Neutralising Antibodies Against HIV-1.Front Immunol. 2021 Oct 19;12:708227. doi: 10.3389/fimmu.2021.708227. eCollection 2021. Front Immunol. 2021. PMID: 34737737 Free PMC article. Review.
-
Broadly neutralizing antibody epitopes on HIV-1 particles are exposed after virus interaction with host cells.J Virol. 2023 Sep 28;97(9):e0071023. doi: 10.1128/jvi.00710-23. Epub 2023 Sep 8. J Virol. 2023. PMID: 37681958 Free PMC article.
-
A Germline-Targeting Chimpanzee SIV Envelope Glycoprotein Elicits a New Class of V2-Apex Directed Cross-Neutralizing Antibodies.mBio. 2023 Feb 28;14(1):e0337022. doi: 10.1128/mbio.03370-22. Epub 2023 Jan 11. mBio. 2023. PMID: 36629414 Free PMC article.
References
-
- Leonard CK, et al. Assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type 1 recombinant human immunodeficiency virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. J. Biol. Chem. 1990;265:10373–10382. - PubMed
-
- Mao Youdong, Wang Liping, Gu Christopher, Herschhorn Alon, Xiang Shi-Hua, Haim Hillel, Yang Xinzhen, Sodroski Joseph. Subunit organization of the membrane-bound HIV-1 envelope glycoprotein trimer. Nature Structural & Molecular Biology. 2012;19(9):893–899. doi: 10.1038/nsmb.2351. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

