Multifunctional sequence-defined macromolecules for chemical data storage
- PMID: 30367037
- PMCID: PMC6203848
- DOI: 10.1038/s41467-018-06926-3
Multifunctional sequence-defined macromolecules for chemical data storage
Abstract
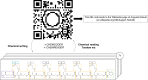
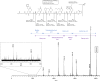
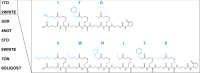
Sequence-defined macromolecules consist of a defined chain length (single mass), end-groups, composition and topology and prove promising in application fields such as anti-counterfeiting, biological mimicking and data storage. Here we show the potential use of multifunctional sequence-defined macromolecules as a storage medium. As a proof-of-principle, we describe how short text fragments (human-readable data) and QR codes (machine-readable data) are encoded as a collection of oligomers and how the original data can be reconstructed. The amide-urethane containing oligomers are generated using an automated protecting-group free, two-step iterative protocol based on thiolactone chemistry. Tandem mass spectrometry techniques have been explored to provide detailed analysis of the oligomer sequences. We have developed the generic software tools Chemcoder for encoding/decoding binary data as a collection of multifunctional macromolecules and Chemreader for reconstructing oligomer sequences from mass spectra to automate the process of chemical writing and reading.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Sequence-Encoded Macromolecules with Increased Data Storage Capacity through a Thiol-Epoxy Reaction.ACS Macro Lett. 2021 May 18;10(5):616-622. doi: 10.1021/acsmacrolett.1c00275. Epub 2021 Apr 30. ACS Macro Lett. 2021. PMID: 35570768
-
Fabrication and Decryption of a Microarray of Digital Dithiosuccinimide Oligomers.Macromol Rapid Commun. 2022 May;43(9):e2200029. doi: 10.1002/marc.202200029. Epub 2022 Apr 1. Macromol Rapid Commun. 2022. PMID: 35322486
-
2D Sequence-Coded Oligourethane Barcodes for Plastic Materials Labeling.Macromol Rapid Commun. 2017 Dec;38(24). doi: 10.1002/marc.201700426. Epub 2017 Aug 21. Macromol Rapid Commun. 2017. PMID: 28833851
-
Reading Information Stored in Synthetic Macromolecules.J Am Chem Soc. 2022 Dec 14;144(49):22378-22390. doi: 10.1021/jacs.2c10316. Epub 2022 Dec 1. J Am Chem Soc. 2022. PMID: 36454647 Review.
-
Reading polymers: sequencing of natural and synthetic macromolecules.Angew Chem Int Ed Engl. 2014 Nov 24;53(48):13010-9. doi: 10.1002/anie.201406766. Epub 2014 Oct 3. Angew Chem Int Ed Engl. 2014. PMID: 25283068 Review.
Cited by
-
Double Porphyrin Cage Compounds.European J Org Chem. 2020 Dec 7;2020(45):7087-7100. doi: 10.1002/ejoc.202001211. Epub 2020 Nov 16. European J Org Chem. 2020. PMID: 33380897 Free PMC article.
-
On-Resin Recycling of Acid-Labile Linker Enables the Reuse of Solid Support for Fmoc-Based Solid Phase Synthesis.Macromol Rapid Commun. 2025 Aug;46(15):e2500073. doi: 10.1002/marc.202500073. Epub 2025 Mar 8. Macromol Rapid Commun. 2025. PMID: 40056078 Free PMC article.
-
From Sequence-Defined Macromolecules to Macromolecular Pin Codes.Adv Sci (Weinh). 2020 Mar 3;7(8):1903698. doi: 10.1002/advs.201903698. eCollection 2020 Apr. Adv Sci (Weinh). 2020. PMID: 32328435 Free PMC article.
-
Secret messaging with endogenous chemistry.Sci Rep. 2021 Jul 6;11(1):13960. doi: 10.1038/s41598-021-92987-2. Sci Rep. 2021. PMID: 34230521 Free PMC article.
-
Enthalpy and entropy synergistic regulation-based programmable DNA motifs for biosensing and information encryption.Sci Adv. 2023 May 19;9(20):eadf5868. doi: 10.1126/sciadv.adf5868. Epub 2023 May 17. Sci Adv. 2023. PMID: 37196083 Free PMC article.
References
-
- Bornholt J, et al. A DNA-based archival storage system. SIGPLAN Not. 2016;51:637–649. doi: 10.1145/2954679.2872397. - DOI
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

