Sexual rejection via a vomeronasal receptor-triggered limbic circuit
- PMID: 30367054
- PMCID: PMC6203846
- DOI: 10.1038/s41467-018-07003-5
Sexual rejection via a vomeronasal receptor-triggered limbic circuit
Abstract
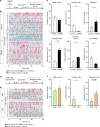
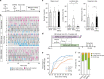
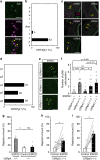
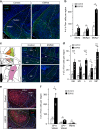
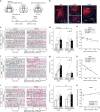
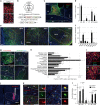
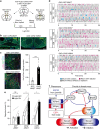
Mating drive is balanced by a need to safeguard resources for offspring, yet the neural basis for negative regulation of mating remains poorly understood. In rodents, pheromones critically regulate sexual behavior. Here, we observe suppression of adult female sexual behavior in mice by exocrine gland-secreting peptide 22 (ESP22), a lacrimal protein from juvenile mice. ESP22 activates a dedicated vomeronasal receptor, V2Rp4, and V2Rp4 knockout eliminates ESP22 effects on sexual behavior. Genetic tracing of ESP22-responsive neural circuits reveals a critical limbic system connection that inhibits reproductive behavior. Furthermore, V2Rp4 counteracts a highly related vomeronasal receptor, V2Rp5, that detects the male sex pheromone ESP1. Interestingly, V2Rp4 and V2Rp5 are encoded by adjacent genes, yet couple to distinct circuits and mediate opposing effects on female sexual behavior. Collectively, our study reveals molecular and neural mechanisms underlying pheromone-mediated sexual rejection, and more generally, how inputs are routed through olfactory circuits to evoke specific behaviors.
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
The male mouse pheromone ESP1 enhances female sexual receptive behaviour through a specific vomeronasal receptor.Nature. 2010 Jul 1;466(7302):118-22. doi: 10.1038/nature09142. Nature. 2010. PMID: 20596023
-
A juvenile mouse pheromone inhibits sexual behaviour through the vomeronasal system.Nature. 2013 Oct 17;502(7471):368-71. doi: 10.1038/nature12579. Epub 2013 Oct 2. Nature. 2013. PMID: 24089208 Free PMC article.
-
Impaired pheromone detection and abnormal sexual behavior in female mice deficient for ancV1R.Curr Biol. 2025 Jan 6;35(1):21-35.e8. doi: 10.1016/j.cub.2024.10.077. Epub 2024 Nov 21. Curr Biol. 2025. PMID: 39577426
-
Molecular biology of peptide pheromone production and reception in mice.Adv Genet. 2007;59:147-71. doi: 10.1016/S0065-2660(07)59006-1. Adv Genet. 2007. PMID: 17888798 Review.
-
Of pheromones and kairomones: what receptors mediate innate emotional responses?Anat Rec (Hoboken). 2013 Sep;296(9):1346-63. doi: 10.1002/ar.22745. Epub 2013 Jul 31. Anat Rec (Hoboken). 2013. PMID: 23904448 Review.
Cited by
-
The vomeronasal organ of wild canids: the fox (Vulpes vulpes) as a model.J Anat. 2020 Nov;237(5):890-906. doi: 10.1111/joa.13254. Epub 2020 Jun 25. J Anat. 2020. PMID: 32584430 Free PMC article.
-
Origin and Evolution of the Gene Family of Proteinaceous Pheromones, the Exocrine Gland-Secreting Peptides, in Rodents.Mol Biol Evol. 2021 Jan 23;38(2):634-649. doi: 10.1093/molbev/msaa220. Mol Biol Evol. 2021. PMID: 32961551 Free PMC article.
-
Coordination of social behaviors by the bed nucleus of the stria terminalis.Eur J Neurosci. 2022 May;55(9-10):2404-2420. doi: 10.1111/ejn.14991. Epub 2020 Nov 7. Eur J Neurosci. 2022. PMID: 33006806 Free PMC article.
-
Bespoke behavior: mechanisms that modulate pheromone-triggered behavior.Curr Opin Neurobiol. 2020 Oct;64:143-150. doi: 10.1016/j.conb.2020.05.003. Epub 2020 Jul 15. Curr Opin Neurobiol. 2020. PMID: 32682209 Free PMC article. Review.
-
A functional cellular framework for sex and estrous cycle-dependent gene expression and behavior.Cell. 2022 Feb 17;185(4):654-671.e22. doi: 10.1016/j.cell.2021.12.031. Epub 2022 Jan 21. Cell. 2022. PMID: 35065713 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

