Using patient-derived iPSCs to develop humanized mouse models for chronic myelomonocytic leukemia and therapeutic drug identification, including liposomal clodronate
- PMID: 30367142
- PMCID: PMC6203784
- DOI: 10.1038/s41598-018-34193-1
Using patient-derived iPSCs to develop humanized mouse models for chronic myelomonocytic leukemia and therapeutic drug identification, including liposomal clodronate
Abstract
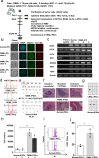
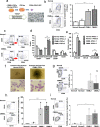
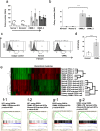
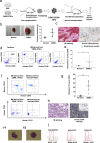
Chronic myelomonocytic leukemia (CMML) is an entity of myelodysplastic syndrome/myeloproliferative neoplasm. Although CMML can be cured with allogeneic stem cell transplantation, its prognosis is generally very poor due to the limited efficacy of chemotherapy and to the patient's age, which is usually not eligible for transplantation. Comprehensive analysis of CMML pathophysiology and the development of therapeutic agents have been limited partly due to the lack of cell lines in CMML and the limited developments of mouse models. After successfully establishing patient's derived disease-specific induced pluripotent stem cells (iPSCs) derived from a patient with CMML, we utilized these CMML-iPSCs to achieve hematopoietic re-differentiation in vitro, created a humanized CMML mouse model via teratomas, and developed a drug-testing system. The clinical characteristics of CMML were recapitulated following hematopoietic re-differentiation in vitro and a humanized CMML mouse model in vivo. The drug-testing system using CMML-iPSCs identified a MEK inhibitor, a Ras inhibitor, and liposomal clodronate as potential drugs for treating CMML. Clodronate is a drug commonly used as a bisphosphonate for osteoporosis. In this study, the liposomalization of clodronate enhanced its effectiveness in these assays, suggesting that this variation of clodronate may be adopted as a repositioned drug for CMML therapy.
Conflict of interest statement
The authors declare no competing interests.
Figures