Organizational principles of 3D genome architecture
- PMID: 30367165
- PMCID: PMC6312108
- DOI: 10.1038/s41576-018-0060-8
Organizational principles of 3D genome architecture
Abstract
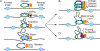
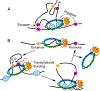
Studies of 3D chromatin organization have suggested that chromosomes are hierarchically organized into large compartments composed of smaller domains called topologically associating domains (TADs). Recent evidence suggests that compartments are smaller than previously thought and that the transcriptional or chromatin state is responsible for interactions leading to the formation of small compartmental domains in all organisms. In vertebrates, CTCF forms loop domains, probably via an extrusion process involving cohesin. CTCF loops cooperate with compartmental domains to establish the 3D organization of the genome. The continuous extrusion of the chromatin fibre by cohesin may also be responsible for the establishment of enhancer-promoter interactions and stochastic aspects of the transcription process. These observations suggest that the 3D organization of the genome is an emergent property of chromatin and its components, and thus may not be only a determinant but also a consequence of its function.
Conflict of interest statement
Competing Interests
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

