OsWRKY67 positively regulates blast and bacteria blight resistance by direct activation of PR genes in rice
- PMID: 30367631
- PMCID: PMC6204034
- DOI: 10.1186/s12870-018-1479-y
OsWRKY67 positively regulates blast and bacteria blight resistance by direct activation of PR genes in rice
Abstract
Background: WRKY proteins are one of the largest gene families and are well-known for their regulatory roles in many aspects of plant development, including plant response to both biotic and abiotic stresses. Although the roles of WRKY proteins in leaf blast resistance have been well-documented in rice, their functions in panicle blast, the most destructive type of blast disease, are still largely unknown.
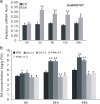
Results: Here, we identified that the transcription of OsWRKY67 was strongly activated by leaf and panicle blast infection. OsWRKY67 is ubiquitously expressed and sub-localized in the nucleus. Rice plants overexpressing OsWRKY67 showed quantitatively enhanced resistance to leaf blast, panicle blast and bacterial blight. In contrast, silencing of OsWRKY67 increased the susceptibility to blast and bacterial blight diseases. RNA-seq analysis indicated that OsWRKY67 induces the transcription of a set of defense-related genes including the ones involved in the salicylic acid (SA)-dependent pathway. Consistent with this, the OsWRKY67-overexpressing plants accumulated higher amounts of endogenous SA, whereas lower endogenous SA levels were observed in OsWRKY67-silenced plants relative to wild-type Nipponbare plants before and after pathogen attack. Moreover, we also observed that OsWRKY67 directly binds to the promoters of PR1a and PR10 to activate their expression.
Conclusions: These results together suggest the positive role of OsWRKY67 in regulating rice responses to leaf blast, panicle blast and bacterial blight disease. Furthermore, conferring resistance to two major diseases makes it a good target of molecular breeding for crop improvement in rice.
Keywords: Bacterial blight; Leaf blast; OsWRKY67; Panicle blast; Salicylic acid.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures






Similar articles
-
The germin-like protein OsGLP2-1 enhances resistance to fungal blast and bacterial blight in rice.Plant Mol Biol. 2016 Nov;92(4-5):411-423. doi: 10.1007/s11103-016-0521-4. Epub 2016 Sep 15. Plant Mol Biol. 2016. PMID: 27631432
-
Overexpression of OsGF14f Enhances Quantitative Leaf Blast and Bacterial Blight Resistance in Rice.Int J Mol Sci. 2022 Jul 4;23(13):7440. doi: 10.3390/ijms23137440. Int J Mol Sci. 2022. PMID: 35806444 Free PMC article.
-
OsGF14b Positively Regulates Panicle Blast Resistance but Negatively Regulates Leaf Blast Resistance in Rice.Mol Plant Microbe Interact. 2016 Jan;29(1):46-56. doi: 10.1094/MPMI-03-15-0047-R. Epub 2015 Dec 10. Mol Plant Microbe Interact. 2016. PMID: 26467468
-
Sheath blight of rice: a review and identification of priorities for future research.Planta. 2019 Nov;250(5):1387-1407. doi: 10.1007/s00425-019-03246-8. Epub 2019 Jul 25. Planta. 2019. PMID: 31346804 Review.
-
Bacterial leaf blight resistance in rice: a review of conventional breeding to molecular approach.Mol Biol Rep. 2019 Feb;46(1):1519-1532. doi: 10.1007/s11033-019-04584-2. Epub 2019 Jan 9. Mol Biol Rep. 2019. PMID: 30628024 Review.
Cited by
-
Every Coin Has Two Sides: Reactive Oxygen Species during Rice⁻Magnaporthe oryzae Interaction.Int J Mol Sci. 2019 Mar 8;20(5):1191. doi: 10.3390/ijms20051191. Int J Mol Sci. 2019. PMID: 30857220 Free PMC article. Review.
-
The OsICS1 is directly regulated by OsWRKY6 and increases resistance against Xanthomonas oryzae pv. oryzae.Planta. 2024 Apr 17;259(6):124. doi: 10.1007/s00425-024-04405-2. Planta. 2024. PMID: 38630137
-
Functional Analysis of Malus hallianaWRKY69 Transcription Factor (TF) Under Iron (Fe) Deficiency Stress.Curr Issues Mol Biol. 2025 Jul 21;47(7):576. doi: 10.3390/cimb47070576. Curr Issues Mol Biol. 2025. PMID: 40729045 Free PMC article.
-
Investigating the Role of OsHDT701 and Other Blast-Associated Negative Regulatory Genes in Indica Rice Cultivar Ranjit Using Combined Wet Lab and Computational Approaches.Mol Biotechnol. 2024 Oct 30. doi: 10.1007/s12033-024-01310-7. Online ahead of print. Mol Biotechnol. 2024. PMID: 39476286
-
OsWRKY70 Plays Opposite Roles in Blast Resistance and Cold Stress Tolerance in Rice.Rice (N Y). 2024 Sep 14;17(1):61. doi: 10.1186/s12284-024-00741-9. Rice (N Y). 2024. PMID: 39271542 Free PMC article.
References
-
- Ou SH, Nuque FL, Bandong JM. Relation between qualitative and quantitative resistance to rice blast. Phytopathology. 1975;65:1315–1316. doi: 10.1094/Phyto-65-1315. - DOI
-
- Parlevliet JE. Components of resistance that reduce the rate of epidemic development. Annu Rev Phytopathol. 1979;1:203–222. doi: 10.1146/annurev.py.17.090179.001223. - DOI
MeSH terms
Substances
Grants and funding
- 31461143019/NSFC-IRRI project
- 2015B020231002/Science and Technology Project of Guangdong Province
- 2016A050502030/Science and Technology Project of Guangdong Province
- 2014B070706013/Science and Technology Project of Guangdong Province
- 2014A030310489/the Natural Science Foundation of Guangdong Province
LinkOut - more resources
Full Text Sources
Research Materials

