A single nucleotide polymorphism in the Plasmodium falciparum atg18 gene associates with artemisinin resistance and confers enhanced parasite survival under nutrient deprivation
- PMID: 30367653
- PMCID: PMC6204056
- DOI: 10.1186/s12936-018-2532-x
A single nucleotide polymorphism in the Plasmodium falciparum atg18 gene associates with artemisinin resistance and confers enhanced parasite survival under nutrient deprivation
Abstract
Background: Artemisinin-resistant Plasmodium falciparum has been reported throughout the Greater Mekong subregion and threatens to disrupt current malaria control efforts worldwide. Polymorphisms in kelch13 have been associated with clinical and in vitro resistance phenotypes; however, several studies suggest that the genetic determinants of resistance may involve multiple genes. Current proposed mechanisms of resistance conferred by polymorphisms in kelch13 hint at a connection to an autophagy-like pathway in P. falciparum.
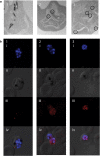
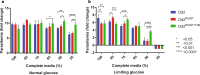
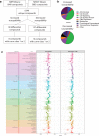
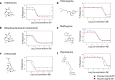
Results: A SNP in autophagy-related gene 18 (atg18) was associated with long parasite clearance half-life in patients following artemisinin-based combination therapy. This gene encodes PfAtg18, which is shown to be similar to the mammalian/yeast homologue WIPI/Atg18 in terms of structure, binding abilities, and ability to form puncta in response to stress. To investigate the contribution of this polymorphism, the atg18 gene was edited using CRISPR/Cas9 to introduce a T38I mutation into a k13-edited Dd2 parasite. The presence of this SNP confers a fitness advantage by enabling parasites to grow faster in nutrient-limited settings. The mutant and parent parasites were screened against drug libraries of 6349 unique compounds. While the SNP did not modulate the parasite's susceptibility to any of the anti-malarial compounds using a 72-h drug pulse, it did alter the parasite's susceptibility to 227 other compounds.
Conclusions: These results suggest that the atg18 T38I polymorphism may provide additional resistance against artemisinin derivatives, but not partner drugs, even in the absence of kelch13 mutations, and may also be important in parasite survival during nutrient deprivation.
Keywords: Artemisinin resistance; Autophagy; Drug resistance; Fitness; atg18.
Figures


References
-
- Duru V, Khim N, Leang R, Kim S, Domergue A, Kloeung N, et al. Plasmodium falciparum dihydroartemisinin-piperaquine failures in Cambodia are associated with mutant K13 parasites presenting high survival rates in novel piperaquine in vitro assays: retrospective and prospective investigations. BMC Med. 2015;13:305. doi: 10.1186/s12916-015-0539-5. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

