Genetic patterns in Neotropical Magnolias (Magnoliaceae) using de novo developed microsatellite markers
- PMID: 30368529
- PMCID: PMC6460770
- DOI: 10.1038/s41437-018-0151-5
Genetic patterns in Neotropical Magnolias (Magnoliaceae) using de novo developed microsatellite markers
Abstract
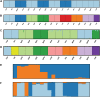
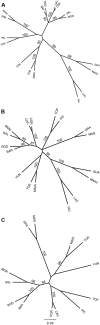
Conserving tree populations safeguards forests since they represent key elements of the ecosystem. The genetic characteristics underlying the evolutionary success of the tree growth form: high genetic diversity, extensive gene flow and strong species integrity, contribute to their survival in terms of adaptability. However, different biological and landscape contexts challenge these characteristics. This study employs 63 de novo developed microsatellite or SSR (Single Sequence Repeat) markers in different datasets of nine Neotropical Magnolia species. The genetic patterns of these protogynous, insect-pollinated tree species occurring in fragmented, highly-disturbed landscapes were investigated. Datasets containing a total of 340 individuals were tested for their genetic structure and degree of inbreeding. Analyses for genetic structure depicted structuring between species, i.e. strong species integrity. Within the species, all but one population pair were considered moderate to highly differentiated, i.e. no indication of extensive gene flow between populations. No overall correlation was observed between genetic and geographic distance of the pairwise species' populations. In contrast to the pronounced genetic structure, there was no evidence of inbreeding within the populations, suggesting mechanisms favouring cross pollination and/or selection for more genetically diverse, heterozygous offspring. In conclusion, the data illustrate that the Neotropical Magnolias in the context of a fragmented landscape still have ample gene flow within populations, yet little gene flow between populations.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


References
-
- Aparicio A, Hampe A, Fernández-Carrillo L, Albaladejo RG. Fragmentation and comparative genetic structure of four mediterranean woody species: complex interactions between life history traits and the landscape context. Divers Distrib. 2012;18(3):226–235.
-
- Brownstein MJ, Carpten JD, Smith JR. Modulation of non-templated nucleotide addition by Taq DNA polymerase: primer modifications that facilitate genotyping. Biotechniques. 1996;20(6):1004–1006. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- K210116N/Fonds Wetenschappelijk Onderzoek (Research Foundation Flanders)/International
- V431816N/Fonds Wetenschappelijk Onderzoek (Research Foundation Flanders)/International
- V445315N/Fonds Wetenschappelijk Onderzoek (Research Foundation Flanders)/International
- BOF.DOC.2014.0062.01/Bijzonder Onderzoeksfonds (Special Research Fund)/International
LinkOut - more resources
Full Text Sources
Other Literature Sources

