Gaining insight into transcriptome-wide RNA population dynamics through the chemistry of 4-thiouridine
- PMID: 30370679
- PMCID: PMC6768404
- DOI: 10.1002/wrna.1513
Gaining insight into transcriptome-wide RNA population dynamics through the chemistry of 4-thiouridine
Abstract
Cellular RNA levels are the result of a juggling act between RNA transcription, processing, and degradation. By tuning one or more of these parameters, cells can rapidly alter the available pool of transcripts in response to stimuli. While RNA sequencing (RNA-seq) is a vital method to quantify RNA levels genome-wide, it is unable to capture the dynamics of different RNA populations at steady-state or distinguish between different mechanisms that induce changes to the steady-state (i.e., altered rate of transcription vs. degradation). The dynamics of different RNA populations can be studied by targeted incorporation of noncanonical nucleosides. 4-Thiouridine (s4 U) is a commonly used and versatile RNA metabolic label that allows the study of many properties of RNA metabolism from synthesis to degradation. Numerous experimental strategies have been developed that leverage the power of s4 U to label newly transcribed RNA in whole cells, followed by enrichment with activated disulfides or chemistry to induce C mutations at sites of s4 U during sequencing. This review presents existing methods to study RNA population dynamics genome-wide using s4 U metabolic labeling, as well as a discussion of considerations and challenges when designing s4 U metabolic labeling experiments. This article is categorized under: RNA Methods > RNA Analyses in Cells RNA Turnover and Surveillance > Regulation of RNA Stability.
Keywords: RNA metabolism; RNA turnover; nucleoside recoding; transcriptome dynamics; transient transcription.
© 2018 Wiley Periodicals, Inc.
Figures

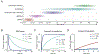
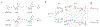


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

