A Transcriptomic Method to Determine Airway Immune Dysfunction in T2-High and T2-Low Asthma
- PMID: 30371106
- PMCID: PMC6376622
- DOI: 10.1164/rccm.201807-1291OC
A Transcriptomic Method to Determine Airway Immune Dysfunction in T2-High and T2-Low Asthma
Abstract
Background: Type 2 (T2) inflammation drives airway dysfunction in many patients with asthma; yet, we lack a comprehensive understanding of the airway immune cell types and networks that sustain this inflammation. Moreover, defects in the airway immune system in patients with asthma without T2 inflammation are not established.
Objectives: To determine the gene networks that sustain T2 airway inflammation in T2-high asthma and to explore the gene networks that characterize T2-low asthma.
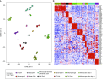
Methods: Network analysis of sputum cell transcriptome expression data from 84 subjects with asthma and 27 healthy control subjects was used to identify immune cell type-enriched networks that underlie asthma subgroups.
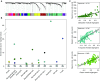
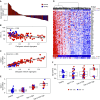
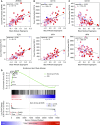
Results: Sputum T2 gene expression was characterized by an immune cell network derived from multiple innate immune cells, including eosinophils, mast cells/basophils, and inflammatory dendritic cells. Clustering of subjects within this network stratified subjects into T2-high and T2-low groups, but it also revealed a subgroup of T2-high subjects with uniformly higher expression of the T2 network. These "T2-ultrahigh subjects" were characterized clinically by older age and more severe airflow obstruction and pathologically by a second T2 network derived from T2-skewed, CD11b+/CD103-/IRF4+ classical dendritic cells. Subjects with T2-low asthma were differentiated from healthy control subjects by lower expression of a cytotoxic CD8+ T-cell network, which was negatively correlated with body mass index and plasma IL-6 concentrations.
Conclusions: Persistent airway T2 inflammation is a complex construct of innate and adaptive immunity gene expression networks that are variable across individuals with asthma and persist despite steroid treatment. Individuals with T2-low asthma exhibit an airway deficiency in cytotoxic T cells associated with obesity-driven inflammation.
Keywords: CD8 cytotoxic T cells; asthma; dendritic cells; sputum gene expression; type 2.
Figures

Comment in
-
Transcriptome Analysis of Sputum Cells. The Modern Art of Assessing Inflammation.Am J Respir Crit Care Med. 2019 Feb 15;199(4):402-404. doi: 10.1164/rccm.201810-2013ED. Am J Respir Crit Care Med. 2019. PMID: 30407844 No abstract available.
References
-
- GBD 2015 Chronic Respiratory Disease Collaborators. Global, regional, and national deaths, prevalence, disability-adjusted life years, and years lived with disability for chronic obstructive pulmonary disease and asthma, 1990-2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet Respir Med. 2017;5:691–706. - PMC - PubMed
-
- Chung KF, Wenzel SE, Brozek JL, Bush A, Castro M, Sterk PJ, et al. International ERS/ATS guidelines on definition, evaluation and treatment of severe asthma. Eur Respir J. 2014;43:343–373. - PubMed
Publication types
MeSH terms
Grants and funding
- K23 HL138303/HL/NHLBI NIH HHS/United States
- P01 HL107202/HL/NHLBI NIH HHS/United States
- R01 HL135156/HL/NHLBI NIH HHS/United States
- P30 DK098722/DK/NIDDK NIH HHS/United States
- U19 AI077439/AI/NIAID NIH HHS/United States
- R01 HL128439/HL/NHLBI NIH HHS/United States
- R01 HL080414/HL/NHLBI NIH HHS/United States
- U10 HL109146/HL/NHLBI NIH HHS/United States
- K12 HL119997/HL/NHLBI NIH HHS/United States
- K24 HL137013/HL/NHLBI NIH HHS/United States
- P01 HL132821/HL/NHLBI NIH HHS/United States
- R01 MD010443/MD/NIMHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

