Ancestral Genomes: a resource for reconstructed ancestral genes and genomes across the tree of life
- PMID: 30371900
- PMCID: PMC6323951
- DOI: 10.1093/nar/gky1009
Ancestral Genomes: a resource for reconstructed ancestral genes and genomes across the tree of life
Abstract
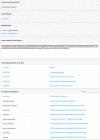
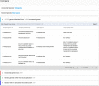
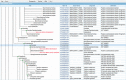
A growing number of whole genome sequencing projects, in combination with development of phylogenetic methods for reconstructing gene evolution, have provided us with a window into genomes that existed millions, and even billions, of years ago. Ancestral Genomes (http://ancestralgenomes.org) is a resource for comprehensive reconstructions of these 'fossil genomes'. Comprehensive sets of protein-coding genes have been reconstructed for 78 genomes of now-extinct species that were the common ancestors of extant species from across the tree of life. The reconstructed genes are based on the extensive library of over 15 000 gene family trees from the PANTHER database, and are updated on a yearly basis. For each ancestral gene, we assign a stable identifier, and provide additional information designed to facilitate analysis: an inferred name, a reconstructed protein sequence, a set of inferred Gene Ontology (GO) annotations, and a 'proxy gene' for each ancestral gene, defined as the least-diverged descendant of the ancestral gene in a given extant genome. On the Ancestral Genomes website, users can browse the Ancestral Genomes by selecting nodes in a species tree, and can compare an extant genome with any of its reconstructed ancestors to understand how the genome evolved.
Figures



References
-
- Fritz-Laylin L.K., Prochnik S.E., Ginger M.L., Dacks J.B., Carpenter M.L., Field M.C., Kuo A., Paredez A., Chapman J., Pham J. et al. . The genome of Naegleria gruberi illuminates early eukaryotic versatility. Cell. 2010; 140:631–642. - PubMed
-
- Doyon J.P., Ranwez V., Daubin V., Berry V.. Models, algorithms and programs for phylogeny reconciliation. Brief Bioinform. 2011; 12:392–400. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

