Identification of Salt Stress Responding Genes Using Transcriptome Analysis in Green Alga Chlamydomonas reinhardtii
- PMID: 30373210
- PMCID: PMC6274750
- DOI: 10.3390/ijms19113359
Identification of Salt Stress Responding Genes Using Transcriptome Analysis in Green Alga Chlamydomonas reinhardtii
Abstract
Salinity is one of the most important abiotic stresses threatening plant growth and agricultural productivity worldwide. In green alga Chlamydomonas reinhardtii, physiological evidence indicates that saline stress increases intracellular peroxide levels and inhibits photosynthetic-electron flow. However, understanding the genetic underpinnings of salt-responding traits in plantae remains a daunting challenge. In this study, the transcriptome analysis of short-term acclimation to salt stress (200 mM NaCl for 24 h) was performed in C. reinhardtii. A total of 10,635 unigenes were identified as being differently expressed by RNA-seq, including 5920 up- and 4715 down-regulated unigenes. A series of molecular cues were screened for salt stress response, including maintaining the lipid homeostasis by regulating phosphatidic acid, acetate being used as an alternative source of energy for solving impairment of photosynthesis, and enhancement of glycolysis metabolism to decrease the carbohydrate accumulation in cells. Our results may help understand the molecular and genetic underpinnings of salt stress responses in green alga C. reinhardtii.
Keywords: Chlamydomonas reinhardtii; impairment of photosynthesis; salt stress; transcriptome analysis; underpinnings of salt stress responses.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
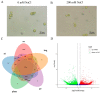

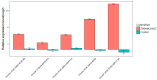
References
-
- Epstein E. Salt-tolerant crops: Origins, development, and prospects of the concept. Plant Soil. 1985;89:187–198. doi: 10.1007/BF02182242. - DOI
-
- Zhang L.Y., Zhang X.J., Fan S.J. Meta-analysis of salt-related gene expression profiles identifies common signatures of salt stress responses in Arabidopsis. Plant Syst. Evol. 2017;303:757–774. doi: 10.1007/s00606-017-1407-x. - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

