Genomic analysis of the Phalaenopsis pathogen Dickeya sp. PA1, representing the emerging species Dickeya fangzhongdai
- PMID: 30373513
- PMCID: PMC6206727
- DOI: 10.1186/s12864-018-5154-3
Genomic analysis of the Phalaenopsis pathogen Dickeya sp. PA1, representing the emerging species Dickeya fangzhongdai
Abstract
Background: Dickeya sp. strain PA1 is the causal agent of bacterial soft rot in Phalaenopsis, an important indoor orchid in China. PA1 and a few other strains were grouped into a novel species, Dickeya fangzhongdai, and only the orchid-associated strains have been shown to cause soft rot symptoms.
Methods: We constructed the complete PA1 genome sequence and used comparative genomics to explore the differences in genomic features between D. fangzhongdai and other Dickeya species.
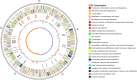
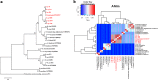
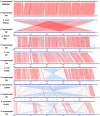
Results: PA1 has a 4,979,223-bp circular genome with 4269 predicted protein-coding genes. D. fangzhongdai was phylogenetically similar to Dickeya solani and Dickeya dadantii. The type I to type VI secretion systems (T1SS-T6SS), except for the stt-type T2SS, were identified in D. fangzhongdai. The three phylogenetically similar species varied significantly in terms of their T5SSs and T6SSs, as did the different D. fangzhongdai strains. Genomic island (GI) prediction and synteny analysis (compared to D. fangzhongdai strains) of PA1 also indicated the presence of T5SSs and T6SSs in strain-specific regions. Two typical CRISPR arrays were identified in D. fangzhongdai and in most other Dickeya species, except for D. solani. CRISPR-1 was present in all of these Dickeya species, while the presence of CRISPR-2 varied due to species differentiation. A large polyketide/nonribosomal peptide (PK/NRP) cluster, similar to the zeamine biosynthetic gene cluster in Dickeya zeae rice strains, was discovered in D. fangzhongdai and D. solani. The D. fangzhongdai and D. solani strains might recently have acquired this gene cluster by horizontal gene transfer (HGT).
Conclusions: Orchid-associated strains are the typical members of D. fangzhongdai. Genomic analysis of PA1 suggested that this strain presents the genomic characteristics of this novel species. Considering the absence of the stt-type T2SS, the presence of CRISPR loci and the zeamine biosynthetic gene cluster, D. fangzhongdai is likely a transitional form between D. dadantii and D. solani. This is supported by the later acquisition of the zeamine cluster and the loss of CRISPR arrays by D. solani. Comparisons of phylogenetic positions and virulence determinants could be helpful for the effective quarantine and control of this emerging species.
Keywords: CRISPR; Comparative genomics; Dickeya; Novel species; PKs/NRPs; Secretion systems.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable. This research did not involve the human subjects, human material, or human data. We isolated the plant pathogen from diseased
Consent for publication
Not applicable.
Competing interests
The authors declare that this research was conducted in the absence of any commercial or financial relationship that could be construed as a potential conflict of interest.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

 flagellar protein and flagellin;
flagellar protein and flagellin;  transcription factor;
transcription factor;  flagellar hook-associated protein;
flagellar hook-associated protein;  flagellar ring protein;
flagellar ring protein;  flagellar motor switch protein;
flagellar motor switch protein;  ATP synthase;
ATP synthase;  flagellar basal-body protein;
flagellar basal-body protein;  methyltransferase;
methyltransferase;  oxidoreductase;
oxidoreductase;  fatty acid synthase;
fatty acid synthase;  transketolase;
transketolase;  acyl carrier protein;
acyl carrier protein;  maltose O-acetyltransferase;
maltose O-acetyltransferase;  aminotransferase;
aminotransferase;  carbamoyl-phosphate synthase;
carbamoyl-phosphate synthase;  transposase;
transposase;  chemotaxis protein;
chemotaxis protein;  chemotaxis family TCS;
chemotaxis family TCS;  flagellar motor protein;
flagellar motor protein;  integrase
integrase
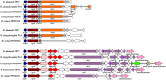


 peroxidase;
peroxidase; 
 ABC transporter;
ABC transporter;  secretion protein HlyD;
secretion protein HlyD;  membrane protein;
membrane protein;  polyketide synthase;
polyketide synthase;  polyunsaturated fatty acid synthase;
polyunsaturated fatty acid synthase;  thioester reductase;
thioester reductase;  hydrolase;
hydrolase;  nonribosomal peptide synthase;
nonribosomal peptide synthase;  phosphopantetheinyl transferase;
phosphopantetheinyl transferase;  hypothetical protein. Circles = the domain predicted in PK or NRP genes. KS = keto reductase domain; AT = acyl transferase domain; KR = keto reductase domain; DH = dehydratase domain; NAD = NAD-binding domain; A = AMP-binding domain; C = condensation domain; E = epimerization domain. The PK domains include KS, AT, KR and DH, and the NRP domains include A, C and E
hypothetical protein. Circles = the domain predicted in PK or NRP genes. KS = keto reductase domain; AT = acyl transferase domain; KR = keto reductase domain; DH = dehydratase domain; NAD = NAD-binding domain; A = AMP-binding domain; C = condensation domain; E = epimerization domain. The PK domains include KS, AT, KR and DH, and the NRP domains include A, C and EReferences
-
- Adeolu M, Alnajar S, Naushad S, Gupta R. Genome-based phylogeny and taxonomy of the ‘Enterobacteriales’: proposal for Enterobacterales Ord. Nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. Nov., Pectobacteriaceae fam. Nov., Yersiniaceae fam. Nov., Hafniaceae fam. Nov., Morganellaceae fam. Nov., and Budviciaceae fam. Nov. Int J Syst Evol Microbiol. 2016;66:5575–5599. doi: 10.1099/ijsem.0.001485. - DOI - PubMed
-
- Samson R, Legendre JB, Christen R, Achouak W, Gardan L. Transfer of Pectobecterium chrysanthemi (Burkholder et al., 1953) Brenner et al. 1973 and Brenneria paradisiaca to the genus Dickeya gen. Nov. as Dickeya chrysanthemi comb. nov. and Dickeya paradisiaca comb. nov. and delineation of four novel species, Dickeya dadantii sp. nov., Dickeya dianthicola sp. nov., Dickeya dieffenbachiae sp. nov. and Dickeya zeae sp. nov. Int J Syst Evol Microbiol. 2005;55:1415–1427. doi: 10.1099/ijs.0.02791-0. - DOI - PubMed
-
- Dye DW, Bradbury JF, Goto M, Hayward AC, Lelliott RA, Schroth MN. International standards for naming pathovars of phytopathogenic bacteria and a list of pathovar names and pathotype strains. Rev of Plant Pathol. 1980;59:153–168.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

