Prediction of pathological complete response and prognosis in patients with neoadjuvant treatment for triple-negative breast cancer
- PMID: 30373556
- PMCID: PMC6206705
- DOI: 10.1186/s12885-018-4925-1
Prediction of pathological complete response and prognosis in patients with neoadjuvant treatment for triple-negative breast cancer
Abstract
Background: It has been reported that pathological complete response is an important surrogate marker for disease-free survival and overall survival in patients with triple-negative breast cancer. This study investigates predictors of the response to neoadjuvant platinum-based or anthracycline-based treatment, and of the prognosis, in patients with triple-negative breast cancer.
Methods: A total of 121 patients with triple-negative breast cancer received neoadjuvant treatment with either platinum or anthracycline between 2008 and 2013. Pathological complete response was assessed relative to different treatments using logistic regression models with age, clinical tumor stage, grading, and Ki-67 as predictors and interaction terms, to obtain adjusted and subgroup-specific results. The impact of the pathological complete response rate on disease-free survival and overall survival was also analyzed.
Results: The pathological complete response rate was higher after platinum/taxane treatment compared with anthracycline/taxane (50.0% vs. 41.8%), but this was not significant in the adjusted analysis (OR 1.44; 95% CI, 0.68 to 3.09). A high histological grade (G3) was a predictor for higher pathological complete response in platinum-based therapy (OR 2.27; 95% CI, 1.00 to 5.30). The effect of neoadjuvant chemotherapy on pathological complete response was significantly different for G1-2 vs. G3 (Pinteraction = 0.013), and additional subgroup-specific differences were noted. Pathological complete response was a predictor for improved disease-free survival and overall survival in both treatment groups, with and without platinum chemotherapy.
Conclusions: This retrospective study of patients with triple-negative breast cancer adds to the evidence that the treatment effect of platinum may be greatest particularly in G3 tumors. In addition, the effect of pathological complete response on the prognosis does not depend on the treatment used.
Keywords: Neoadjuvant therapy; Pathological complete response; Platinum; Prediction; Prognosis; Triple-negative breast cancer.
Conflict of interest statement
Ethics approval and consent to participate
This retrospective study and the anonymized scientific use of the data were approved by the ethics committee of the medical faculty of the University of Erlangen–Nuremberg. Informed consent was obtained from each individual participant included in the study.
Consent for publication
Not applicable.
Competing interests
PG has received honoraria from Novartis and financial support for symposia from Novartis, Roche, and PharmaMar. MPL has received honoraria from MSD and AstraZeneca. A Hartmann is member of the Editorial Board of BMC Cancer as Associate Editor. PAF has received honoraria from Roche, Amgen, Celgene, Novartis, and Pfizer. The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

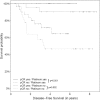
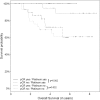
References
-
- Robert-Koch-Institut . Krebs in Deutschland für 2013/2014. 2017.
-
- Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98(19):10869–10874. doi: 10.1073/pnas.191367098. - DOI - PMC - PubMed
-
- Sotiriou C, Neo SY, McShane LM, Korn EL, Long PM, Jazaeri A, Martiat P, Fox SB, Harris AL, Liu ET. Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc Natl Acad Sci U S A. 2003;100(18):10393–10398. doi: 10.1073/pnas.1732912100. - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

