Non-specificity of sequence characterised amplified region as an alternative molecular epidemiology marker for the identification of Salmonella enterica subspecies enterica serovar Typhi
- PMID: 30373642
- PMCID: PMC6206845
- DOI: 10.1186/s13104-018-3870-z
Non-specificity of sequence characterised amplified region as an alternative molecular epidemiology marker for the identification of Salmonella enterica subspecies enterica serovar Typhi
Abstract
Objective: Identification of Salmonella Typhi by conventional culture techniques is labour-intensive, time consuming, and lack sensitivity and specificity unlike high-throughput epidemiological markers that are highly specific but are not affordable for low-resource settings. SCAR, obtained from RAPD technique, is an affordable, reliable and reproducible method for developing genetic markers. Hence, this study investigated the use of SCAR as an alternative molecular epidemiological marker for easy identification of S. Typhi in low-resource settings.
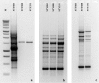
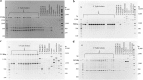
Results: One hundred and twenty RAPD primers were screened through RAPD-PCR against a panel of common enterobacteriaceae for the best RAPD band pattern discrimination to develop SCAR primers that were used to develop a RAPD-SCAR PCR. Of this number, 10 were selected based on their calculated indices of discrimination. Four RAPD primers, SBSA02, SBSA03, SBSD08 and SBSD11 produced suitable bands ranging from 900 to 2500 bp. However, only SBSD11 was found to be specific for S. Typhi, and was cloned, sequenced and used to design new SCAR primers. The primers were used to amplify a panel of organisms to evaluate its specificity. However, the amplified regions were similar to other non-Typhi genomes denoting a lack of specificity of the primers as a marker for S. Typhi.
Keywords: Kelantan; Malaysia; RAPD; S. Typhi; SCAR.
Figures

Similar articles
-
Optimization of randomly amplified polymorphic DNA-polymerase chain reaction for molecular typing of Salmonella enterica serovar Typhi.Rev Soc Bras Med Trop. 2004 Mar-Apr;37(2):143-7. doi: 10.1590/s0037-86822004000200006. Epub 2004 Apr 13. Rev Soc Bras Med Trop. 2004. PMID: 15094899
-
Application of random amplified polymorphic DNA analysis to differentiate strains of Salmonella typhi and other Salmonella species.J Appl Microbiol. 1998 Oct;85(4):693-702. doi: 10.1111/j.1365-2672.1998.00582.x. J Appl Microbiol. 1998. PMID: 9812381
-
Molecular typing of Salmonella enterica serovar typhi isolates from various countries in Asia by a multiplex PCR assay on variable-number tandem repeats.J Clin Microbiol. 2003 Sep;41(9):4388-94. doi: 10.1128/JCM.41.9.4388-4394.2003. J Clin Microbiol. 2003. PMID: 12958274 Free PMC article.
-
Use of TaqMan® real-time PCR for rapid detection of Salmonella enterica serovar Typhi.Acta Microbiol Immunol Hung. 2014 Jun;61(2):121-30. doi: 10.1556/AMicr.61.2014.2.3. Acta Microbiol Immunol Hung. 2014. PMID: 24939681
-
Application of randomly amplified polymorphic DNA (RAPD) analysis for typing avian Salmonella enterica subsp. enterica.FEMS Immunol Med Microbiol. 2000 Nov;29(3):221-5. doi: 10.1111/j.1574-695X.2000.tb01526.x. FEMS Immunol Med Microbiol. 2000. PMID: 11064269
Cited by
-
Effectiveness of six molecular typing methods as epidemiological tools for the study of Salmonella isolates in two Colombian regions.Vet World. 2019 Dec;12(12):1998-2006. doi: 10.14202/vetworld.2019.1998-2006. Epub 2019 Dec 19. Vet World. 2019. PMID: 32095053 Free PMC article.
-
Comparative Analysis Between Paramecium Strains with Different Syngens Using the RAPD Method.Microb Ecol. 2022 Aug;84(2):594-602. doi: 10.1007/s00248-021-01864-y. Epub 2021 Sep 15. Microb Ecol. 2022. PMID: 34522990
References
-
- Jin U-H, Cho S-H, Kim M-G, Ha S-D, Kim K-S, Lee K-H, Kim K-Y, Chung DH, Lee Y-C, Kim C-H. PCR method based on the ogdH gene for the detection of Salmonella spp. from chicken meat samples. J Microbiol. 2004;42(3):216–222. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

