A nucleosome-free region locally abrogates histone H1-dependent restriction of linker DNA accessibility in chromatin
- PMID: 30373774
- PMCID: PMC6302178
- DOI: 10.1074/jbc.RA118.005721
A nucleosome-free region locally abrogates histone H1-dependent restriction of linker DNA accessibility in chromatin
Abstract
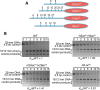
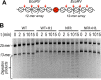
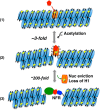
Eukaryotic genomes are packaged into linker-oligonucleosome assemblies, providing compaction of genomic DNA and contributing to gene regulation and genome integrity. To define minimal requirements for initial steps in the transition of compact, closed chromatin to a transcriptionally active, open state, we developed a model in vitro system containing a single, unique, "target" nucleosome in the center of a 25-nucleosome array and evaluated the accessibility of the linker DNA adjacent to this target nucleosome. We found that condensation of H1-lacking chromatin results in ∼60-fold reduction in linker DNA accessibility and that mimics of acetylation within all four core histone tail domains of the target nucleosome synergize to increase accessibility ∼3-fold. Notably, stoichiometric binding of histone H1 caused >2 orders of magnitude reduction in accessibility that was marginally diminished by histone acetylation mimics. Remarkably, a nucleosome-free region (NFR) in place of the target nucleosome completely abrogated H1-dependent restriction of linker accessibility in the immediate vicinity of the NFR. Our results suggest that linker DNA is as inaccessible as DNA within the nucleosome core in fully condensed, H1-containing chromatin. They further imply that an unrecognized function of NFRs in gene promoter regions is to locally abrogate the severe restriction of linker DNA accessibility imposed by H1s.
Keywords: chromatin; chromatin regulation; chromatin structure; epigenetics; gene regulation; histone; histone H1; histone acetylation; linker histone; nucleosome; nucleosome free region.
© 2018 Mishra and Hayes.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures



References
-
- Maeshima K., Rogge R., Tamura S., Joti Y., Hikima T., Szerlong H., Krause C., Herman J., Seidel E., DeLuca J., Ishikawa T., and Hansen J. C. (2016) Nucleosomal arrays self-assemble into supramolecular globular structures lacking 30-nm fibers. EMBO J. 35, 1115–1132 10.15252/embj.201592660 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

