Increased Mitochondrial Biogenesis and Reactive Oxygen Species Production Accompany Prolonged CD4+ T Cell Activation
- PMID: 30373851
- PMCID: PMC6246812
- DOI: 10.4049/jimmunol.1800753
Increased Mitochondrial Biogenesis and Reactive Oxygen Species Production Accompany Prolonged CD4+ T Cell Activation
Abstract
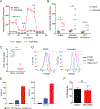
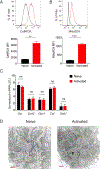
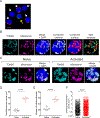
Activation of CD4+ T cells to proliferate drives cells toward aerobic glycolysis for energy production while using mitochondria primarily for macromolecular synthesis. In addition, the mitochondria of activated T cells increase production of reactive oxygen species, providing an important second messenger for intracellular signaling pathways. To better understand the critical changes in mitochondria that accompany prolonged T cell activation, we carried out an extensive analysis of mitochondrial remodeling using a combination of conventional strategies and a novel high-resolution imaging method. We show that for 4 d following activation, mouse CD4+ T cells sustained their commitment to glycolysis facilitated by increased glucose uptake through increased expression of GLUT transporters. Despite their limited contribution to energy production, mitochondria were active and showed increased reactive oxygen species production. Moreover, prolonged activation of CD4+ T cells led to increases in mitochondrial content and volume, in the number of mitochondria per cell and in mitochondrial biogenesis. Thus, during prolonged activation, CD4+ T cells continue to obtain energy predominantly from glycolysis but also undergo extensive mitochondrial remodeling, resulting in increased mitochondrial activity.
Conflict of interest statement
Authors declare no conflicts of interest.
Figures




References
-
- Buck MD, O’Sullivan D, Klein Geltink RI, Curtis JD, Chang CH, Sanin DE, Qiu J, Kretz O, Braas D, van der Windt GJ, Chen Q, Huang SC, O’Neill CM, Edelson BT, Pearce EJ, Sesaki H, Huber TB, Rambold AS, and Pearce EL. 2016. Mitochondrial Dynamics Controls T Cell Fate through Metabolic Programming. Cell 166: 63–76. - PMC - PubMed
-
- Caro-Maldonado A, Wang R, Nichols AG, Kuraoka M, Milasta S, Sun LD, Gavin AL, Abel ED, Kelsoe G, Green DR, and Rathmell JC. 2014. Metabolic reprogramming is required for antibody production that is suppressed in anergic but exaggerated in chronically BAFF-exposed B cells. J Immunol 192: 3626–3636. - PMC - PubMed
-
- Frauwirth KA, Riley JL, Harris MH, Parry RV, Rathmell JC, Plas DR, Elstrom RL, June CH, and Thompson CB. 2002. The CD28 signaling pathway regulates glucose metabolism. Immunity 16: 769–777. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

