Homeostasis of protein and mRNA concentrations in growing cells
- PMID: 30374016
- PMCID: PMC6206055
- DOI: 10.1038/s41467-018-06714-z
Homeostasis of protein and mRNA concentrations in growing cells
Abstract
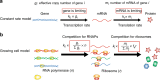
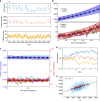
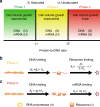
Many experiments show that the numbers of mRNA and protein are proportional to the cell volume in growing cells. However, models of stochastic gene expression often assume constant transcription rate per gene and constant translation rate per mRNA, which are incompatible with these experiments. Here, we construct a minimal gene expression model to fill this gap. Assuming ribosomes and RNA polymerases are limiting in gene expression, we show that the numbers of proteins and mRNAs both grow exponentially during the cell cycle and that the concentrations of all mRNAs and proteins achieve cellular homeostasis; the competition between genes for the RNA polymerases makes the transcription rate independent of the genome number. Furthermore, by extending the model to situations in which DNA (mRNA) can be saturated by RNA polymerases (ribosomes) and becomes limiting, we predict a transition from exponential to linear growth of cell volume as the protein-to-DNA ratio increases.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Paulsson J. Models of stochastic gene expression. Phys. Life Rev. 2005;2:157–175. doi: 10.1016/j.plrev.2005.03.003. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

