Comprehensive Longitudinal Microbiome Analysis of the Chicken Cecum Reveals a Shift From Competitive to Environmental Drivers and a Window of Opportunity for Campylobacter
- PMID: 30374341
- PMCID: PMC6196313
- DOI: 10.3389/fmicb.2018.02452
Comprehensive Longitudinal Microbiome Analysis of the Chicken Cecum Reveals a Shift From Competitive to Environmental Drivers and a Window of Opportunity for Campylobacter
Abstract
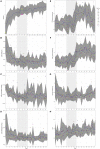
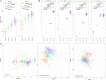
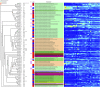
Chickens are a key food source for humans yet their microbiome contains bacteria that can be pathogenic to humans, and indeed potentially to chickens themselves. Campylobacter is present within the chicken gut and is the leading cause of bacterial foodborne gastroenteritis within humans worldwide. Infection can lead to secondary sequelae such as Guillain-Barré syndrome and stunted growth in children from low-resource areas. Despite the global health impact and economic burden of Campylobacter, how and when Campylobacter appears within chickens remains unclear. The lack of day to day microbiome data with replicates, relevant metadata, and a lack of natural infection studies have delayed our understanding of the chicken gut microbiome and Campylobacter. Here, we performed a comprehensive day to day microbiome analysis of the chicken cecum from day 3 to 35 (12 replicates each day; final n = 379). We combined metadata such as chicken weight and feed conversion rates to investigate what the driving forces are for the microbial changes within the chicken gut over time, and how this relates to Campylobacter appearance within a natural habitat setting. We found a rapidly increasing microbial diversity up to day 12 with variation observed both in terms of genera and abundance, before a stabilization of the microbial diversity after day 20. In particular, we identified a shift from competitive to environmental drivers of microbial community from days 12 to 20 creating a window of opportunity whereby Campylobacter can appear. Campylobacter was identified at day 16 which was 1 day after the most substantial changes in metabolic profiles observed. In addition, microbial variation over time is most likely influenced by the diet of the chickens whereby significant shifts in OTU abundances and beta dispersion of samples often corresponded with changes in feed. This study is unique in comparison to the most recent studies as neither sampling was sporadic nor Campylobacter was artificially introduced, thus the experiments were performed in a natural setting. We believe that our findings can be useful for future intervention strategies and help reduce the burden of Campylobacter within the food chain.
Keywords: Campylobacter; chicken; competitive exclusion; diversity; environmental filtering; microbiome; phylogenetic signal.
Figures
References
-
- Apajalahti J. (2005). Comparative gut microflora, metabolic challenges, and potential opportunities. J. Appl. Poult. Res. 14, 444–453. 10.1093/japr/14.2.444 - DOI
-
- Asshauer K. P., Meinicke P. (2013). On the estimation of metabolic profiles in metagenomics, in German Conference on Bioinformatics 2013, eds Beissbarth T., Kollmar M., Leha A., Morgenstern B., Schultz A., Waack S., Wingender E. (Dagstuhl: Schloss Dagstuhl–Leibniz-Zentrum fuer Informatik; ), 1–13.
LinkOut - more resources
Full Text Sources
Other Literature Sources

