Subdiffusion of loci and cytoplasmic particles are different in compressed Escherichia coli cells
- PMID: 30374466
- PMCID: PMC6200837
- DOI: 10.1038/s42003-018-0185-5
Subdiffusion of loci and cytoplasmic particles are different in compressed Escherichia coli cells
Abstract
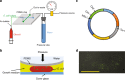
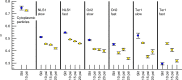
The complex physical nature of the bacterial intracellular environment remains largely unknown, and has relevance for key biochemical and biological processes of the cell. Although recent work has addressed the role of non-equilibrium sources of activity and crowding, the consequences of mechanical perturbations are relatively less explored. Here we use a microfabricated valve system to track both fluorescently labeled chromosomal loci and cytoplasmic particles in Escherichia coli cells shortly after applying a compressive force, observing the response on time scales that are too sudden to allow for biochemical response from the cell. Cytoplasmic diffusion slows markedly on compression but the exponent governing the growth of the ensemble-averaged mean-squared displacement of cytoplasmic particles is unaffected. In contrast, the corresponding exponent for DNA loci changes significantly. These results suggest that DNA elasticity and nucleoid organization play a more important role in loci subdiffusion than cytoplasmic viscoelasticity under such short time scales.
Conflict of interest statement
The authors declare no competing Interests.
Figures


Similar articles
-
Mesoscale Simulation of Bacterial Chromosome and Cytoplasmic Nanoparticles in Confinement.Entropy (Basel). 2021 Apr 28;23(5):542. doi: 10.3390/e23050542. Entropy (Basel). 2021. PMID: 33924872 Free PMC article.
-
Bacterial chromosomal loci move subdiffusively through a viscoelastic cytoplasm.Phys Rev Lett. 2010 Jun 11;104(23):238102. doi: 10.1103/PhysRevLett.104.238102. Epub 2010 Jun 8. Phys Rev Lett. 2010. PMID: 20867274 Free PMC article.
-
The effects of polydisperse crowders on the compaction of the Escherichia coli nucleoid.Mol Microbiol. 2020 May;113(5):1022-1037. doi: 10.1111/mmi.14467. Epub 2020 Feb 5. Mol Microbiol. 2020. PMID: 31961016 Free PMC article.
-
Structural and physical aspects of bacterial chromosome segregation.J Struct Biol. 2006 Nov;156(2):273-83. doi: 10.1016/j.jsb.2006.04.013. Epub 2006 May 20. J Struct Biol. 2006. PMID: 16828313 Review.
-
Wanted: a positive control for anomalous subdiffusion.Biophys J. 2012 Dec 19;103(12):2411-22. doi: 10.1016/j.bpj.2012.10.038. Epub 2012 Dec 18. Biophys J. 2012. PMID: 23260043 Free PMC article. Review.
Cited by
-
Dynamics of Proteins and Macromolecular Machines in Escherichia coli.EcoSal Plus. 2021 Dec 15;9(2):eESP00112020. doi: 10.1128/ecosalplus.ESP-0011-2020. Epub 2021 Jun 1. EcoSal Plus. 2021. PMID: 34060908 Free PMC article. Review.
-
Microfluidic techniques for mechanical measurements of biological samples.Biophys Rev (Melville). 2023 Jan 20;4(1):011303. doi: 10.1063/5.0130762. eCollection 2023 Mar. Biophys Rev (Melville). 2023. PMID: 38505816 Free PMC article. Review.
-
A Novel Fractional Brownian Dynamics Method for Simulating the Dynamics of Confined Bottle-Brush Polymers in Viscoelastic Solution.Polymers (Basel). 2024 Feb 15;16(4):524. doi: 10.3390/polym16040524. Polymers (Basel). 2024. PMID: 38399901 Free PMC article.
-
Fluctuating Diffusivity of RNA-Protein Particles: Analogy with Thermodynamics.Entropy (Basel). 2021 Mar 12;23(3):333. doi: 10.3390/e23030333. Entropy (Basel). 2021. PMID: 33808996 Free PMC article.
-
Mesoscale Simulation of Bacterial Chromosome and Cytoplasmic Nanoparticles in Confinement.Entropy (Basel). 2021 Apr 28;23(5):542. doi: 10.3390/e23050542. Entropy (Basel). 2021. PMID: 33924872 Free PMC article.

