Use of a Fluorescent Analogue of a HBV Core Protein-Directed Drug To Interrogate an Antiviral Mechanism
- PMID: 30375863
- PMCID: PMC6350927
- DOI: 10.1021/jacs.8b07988
Use of a Fluorescent Analogue of a HBV Core Protein-Directed Drug To Interrogate an Antiviral Mechanism
Abstract
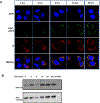
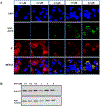
Heteroaryldihydropyrimidines (HAPs) are antiviral small molecules that enhance assembly of HBV core protein (Cp), lead to assembly of empty and defective particles, and suppress viral replication. These core protein allosteric modulators (CpAMs) bind to the pocket at the interface between two Cp dimers and strengthen interdimer interactions. To investigate the CpAM mechanism, we wanted to examine the cellular distributions of Cp and the CpAM itself. For this reason, we developed a fluorescently labeled CpAM, HAP-ALEX. In vitro, HAP-ALEX modulated assembly of purified Cp and at saturating concentrations induced formation of large structures. HAP-ALEX bound capsids and not dimers, making it a capsid-specific molecular tag. HAP-ALEX labeled HBV in transfected cells, with no detectable background with a HAP-insensitive Cp mutant. HAP-ALEX caused redistribution of Cp in a dose-dependent manner consistent with its 0.7 μM EC50, leading to formation of large puncta and an exclusively cytoplasmic distribution. HAP-ALEX colocalized with the redistributed Cp, but large puncta accumulated long before they appeared saturated with the fluorescent CpAM. CpAMs affect HBV assembly and localization; with a fluorescent CpAM both drug and target can be identified.
Figures
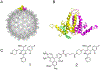




References
-
- Lavanchy D Hepatitis B virus epidemiology, disease burden, treatment, and current and emerging prevention and control measures. J Viral Hepat 2004, 11, 97. - PubMed
-
- Ott JJ; Stevens GA; Groeger J; Wiersma ST Global epidemiology of hepatitis B virus infection: new estimates of age-specific HBsAg seroprevalence and endemicity. Vaccine 2012, 30, 2212. - PubMed
-
- Lozano R; Naghavi M; Foreman K; Lim S; Shibuya K; Aboyans V; Abraham J; Adair T; Aggarwal R; Ahn SY; et, a. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 2012, 380, 2095. - PMC - PubMed
-
- Buti M HBeAg-positive chronic hepatitis B: why do i treat my patients with Nucleos(t)ide analogs? Liver Int 2014, 34 Suppl 1, 108. - PubMed
-
- Jordheim LP; Durantel D; Zoulim F; Dumontet C Advances in the development of nucleoside and nucleotide analogues for cancer and viral diseases. Nat Rev Drug Discov 2013, 12, 447. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

