Proficiency Testing of Standardized Samples Shows Very High Interlaboratory Agreement for Clinical Next-Generation Sequencing-Based Oncology Assays
- PMID: 30376374
- PMCID: PMC6910717
- DOI: 10.5858/arpa.2018-0336-CP
Proficiency Testing of Standardized Samples Shows Very High Interlaboratory Agreement for Clinical Next-Generation Sequencing-Based Oncology Assays
Abstract
Context.—: Next-generation sequencing-based assays are being increasingly used in the clinical setting for the detection of somatic variants in solid tumors, but limited data are available regarding the interlaboratory performance of these assays.
Objective.—: To examine proficiency testing data from the initial College of American Pathologists (CAP) Next-Generation Sequencing Solid Tumor survey to report on laboratory performance.
Design.—: CAP proficiency testing results from 111 laboratories were analyzed for accuracy and associated assay performance characteristics.
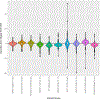
Results.—: The overall accuracy observed for all variants was 98.3%. Rare false-negative results could not be attributed to sequencing platform, selection method, or other assay characteristics. The median and average of the variant allele fractions reported by the laboratories were within 10% of those orthogonally determined by digital polymerase chain reaction for each variant. The median coverage reported at the variant sites ranged from 1922 to 3297.
Conclusions.—: Laboratories demonstrated an overall accuracy of greater than 98% with high specificity when examining 10 clinically relevant somatic single-nucleotide variants with a variant allele fraction of 15% or greater. These initial data suggest excellent performance, but further ongoing studies are needed to evaluate the performance of lower variant allele fractions and additional variant types.
Figures


References
-
- Allen NE, Sudlow C, Peakman T, Collins R. UK biobank data: come and get it. Sci Transl Med 2014;6(224):224ed224. - PubMed
-
- Laverty H, Goldman M. Editorial: The Innovative Medicines Initiative--collaborations are key to innovating R&D processes. Biotechnol J 2014;9(9):1095–1096. - PubMed
-
- Singer DS, Jacks T, Jaffee E. A U.S. “Cancer Moonshot” to accelerate cancer research. Science 2016;353(6304):1105–1106. - PubMed
-
- Bombard Y, Bach PB, Offit K. Translating genomics in cancer care. J Natl Compr Canc Netw 2013;11(11):1343–1353. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
