QTL mapping of natural variation reveals that the developmental regulator bruno reduces tolerance to P-element transposition in the Drosophila female germline
- PMID: 30376574
- PMCID: PMC6207299
- DOI: 10.1371/journal.pbio.2006040
QTL mapping of natural variation reveals that the developmental regulator bruno reduces tolerance to P-element transposition in the Drosophila female germline
Abstract
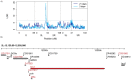
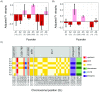
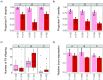
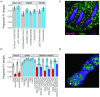
Transposable elements (TEs) are obligate genetic parasites that propagate in host genomes by replicating in germline nuclei, thereby ensuring transmission to offspring. This selfish replication not only produces deleterious mutations-in extreme cases, TE mobilization induces genotoxic stress that prohibits the production of viable gametes. Host genomes could reduce these fitness effects in two ways: resistance and tolerance. Resistance to TE propagation is enacted by germline-specific small-RNA-mediated silencing pathways, such as the Piwi-interacting RNA (piRNA) pathway, and is studied extensively. However, it remains entirely unknown whether host genomes may also evolve tolerance by desensitizing gametogenesis to the harmful effects of TEs. In part, the absence of research on tolerance reflects a lack of opportunity, as small-RNA-mediated silencing evolves rapidly after a new TE invades, thereby masking existing variation in tolerance. We have exploited the recent historical invasion of the Drosophila melanogaster genome by P-element DNA transposons in order to study tolerance of TE activity. In the absence of piRNA-mediated silencing, the genotoxic stress imposed by P-elements disrupts oogenesis and, in extreme cases, leads to atrophied ovaries that completely lack germline cells. By performing quantitative trait locus (QTL) mapping on a panel of recombinant inbred lines (RILs) that lack piRNA-mediated silencing of P-elements, we uncovered multiple QTL that are associated with differences in tolerance of oogenesis to P-element transposition. We localized the most significant QTL to a small 230-kb euchromatic region, with the logarithm of the odds (LOD) peak occurring in the bruno locus, which codes for a critical and well-studied developmental regulator of oogenesis. Genetic, cytological, and expression analyses suggest that bruno dosage modulates germline stem cell (GSC) loss in the presence of P-element activity. Our observations reveal segregating variation in TE tolerance for the first time, and implicate gametogenic regulators as a source of tolerant variants in natural populations.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Comment in
-
Invasion of the P elements: Tolerance is not futile.PLoS Biol. 2018 Oct 30;16(10):e3000036. doi: 10.1371/journal.pbio.3000036. eCollection 2018 Oct. PLoS Biol. 2018. PMID: 30376563 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

