Selective sweeps and genetic lineages of Plasmodium falciparum multi-drug resistance (pfmdr1) gene in Kenya
- PMID: 30376843
- PMCID: PMC6208105
- DOI: 10.1186/s12936-018-2534-8
Selective sweeps and genetic lineages of Plasmodium falciparum multi-drug resistance (pfmdr1) gene in Kenya
Abstract
Background: There are concerns that resistance to artemisinin-based combination therapy might emerge in Kenya and sub-Saharan Africa (SSA) in the same pattern as was with chloroquine and sulfadoxine-pyrimethamine. Single nucleotide polymorphisms (SNPs) in critical alleles of pfmdr1 gene have been associated with resistance to artemisinin and its partner drugs. Microsatellite analysis of loci flanking genes associated with anti-malarial drug resistance has been used in defining the geographic origins, dissemination of resistant parasites and identifying regions in the genome that have been under selection.
Methods: This study set out to investigate evidence of selective sweep and genetic lineages in pfmdr1 genotypes associated with the use of artemether-lumefantrine (AL), as the first-line treatment in Kenya. Parasites (n = 252) from different regions in Kenya were assayed for SNPs at codons 86, 184 and 1246 and typed for 7 neutral microsatellites and 13 microsatellites loci flanking (± 99 kb) pfmdr1 in Plasmodium falciparum infections.
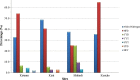
Results: The data showed differential site and region specific prevalence of SNPs associated with drug resistance in the pfmdr1 gene. The prevalence of pfmdr1 N86, 184F, and D1246 in western Kenya (Kisumu, Kericho and Kisii) compared to the coast of Kenya (Malindi) was 92.9% vs. 66.7%, 53.5% vs. to 24.2% and 96% vs. to 87.9%, respectively. The NFD haplotype which is consistent with AL selection was at 51% in western Kenya compared to 25% in coastal Kenya.
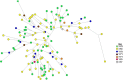
Conclusion: Selection pressures were observed to be different in different regions of Kenya, especially the western region compared to the coastal region. The data showed independent genetic lineages for all the pfmdr1 alleles. The evidence of soft sweeps in pfmdr1 observed varied in direction from one region to another. This is challenging for malaria control programs in SSA which clearly indicate effective malaria control policies should be based on the region and not at a country wide level.
Keywords: Artemisinin resistance; Genetic lineages; Soft selective sweeps.
Figures

References
-
- WHO . World malaria report 2009. Geneva: World Health Organization; 2009.
-
- WHO . World malaria report 2014. Geneva: World Health Organization; 2015.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

