ATF3 inhibits the tumorigenesis and progression of hepatocellular carcinoma cells via upregulation of CYR61 expression
- PMID: 30376856
- PMCID: PMC6208028
- DOI: 10.1186/s13046-018-0919-8
ATF3 inhibits the tumorigenesis and progression of hepatocellular carcinoma cells via upregulation of CYR61 expression
Abstract
Background: Hepatocellular carcinoma (HCC) is one of the most common malignant cancers with a high incidence and high mortality in East Asia. Identifying biomarkers and clarifying the regulatory mechanisms of HCC are of great importance. Herein, we report the role and mechanism of activating transcription factor 3 (ATF3), a member of the ATF/cAMP-responsive element-binding protein family of transcription factors in HCC.
Methods: ATF3 overexpression vector and shRNAs were transfected into HCC cancer cells to upregulate or downregulate ATF3 expression. In vitro and in vivo assays were performed to investigate the functional role of ATF3 in hepatocellular carcinoma. RNA-Seq was performed to screen the differentially expressed genes downstream of ATF3. The dual-luciferase reporter assay, chromatin immunoprecipitation (Ch-IP) analysis and functional rescue experiments were used to confirm the target gene regulated by ATF3. Tissue microarrays (TMAs) comprising 236 human primary HCC tissues were obtained and immunohistochemical staining were carried out to analyze the clinical significance of ATF3.
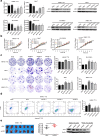
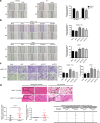
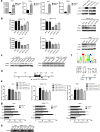
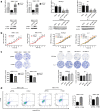
Results: The results indicate that ATF3 significantly inhibited the proliferation and mobility of HCC cells both in vitro and in vivo. Cysteine-rich angiogenic inducer 61 (CYR61) is a key target for transcriptional regulation by ATF3. Both ATF3 and CYR61 were consistently downregulated in human HCC tissues, and their expression levels were significantly and positively correlated with each other.
Conclusions: Our findings indicate that ATF3 functions as a tumor suppressor in HCC through targeting and regulating CYR61.
Keywords: ATF3; CYR61; Hepatocellular carcinoma.
Conflict of interest statement
Ethics approval and consent to participate
All animal procedures were performed under the guidelines of the Shanghai Medical Experimental Animal Care Commission. The study was approved by the Chinese Ethical Review Committee and signed informed consent was obtained from each patient.
Consent for publication
All the patients that involved in the study have given their consent to publish their individual data.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

