Distinct microbes, metabolites, and ecologies define the microbiome in deficient and proficient mismatch repair colorectal cancers
- PMID: 30376889
- PMCID: PMC6208080
- DOI: 10.1186/s13073-018-0586-6
Distinct microbes, metabolites, and ecologies define the microbiome in deficient and proficient mismatch repair colorectal cancers
Abstract
Background: Links between colorectal cancer (CRC) and the gut microbiome have been established, but the specific microbial species and their role in carcinogenesis remain an active area of inquiry. Our understanding would be enhanced by better accounting for tumor subtype, microbial community interactions, metabolism, and ecology.
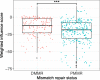
Methods: We collected paired colon tumor and normal-adjacent tissue and mucosa samples from 83 individuals who underwent partial or total colectomies for CRC. Mismatch repair (MMR) status was determined in each tumor sample and classified as either deficient MMR (dMMR) or proficient MMR (pMMR) tumor subtypes. Samples underwent 16S rRNA gene sequencing and a subset of samples from 50 individuals were submitted for targeted metabolomic analysis to quantify amino acids and short-chain fatty acids. A PERMANOVA was used to identify the biological variables that explained variance within the microbial communities. dMMR and pMMR microbial communities were then analyzed separately using a generalized linear mixed effects model that accounted for MMR status, sample location, intra-subject variability, and read depth. Genome-scale metabolic models were then used to generate microbial interaction networks for dMMR and pMMR microbial communities. We assessed global network properties as well as the metabolic influence of each microbe within the dMMR and pMMR networks.
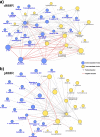
Results: We demonstrate distinct roles for microbes in dMMR and pMMR CRC. Bacteroides fragilis and sulfidogenic Fusobacterium nucleatum were significantly enriched in dMMR CRC, but not pMMR CRC. These findings were further supported by metabolic modeling and metabolomics indicating suppression of B. fragilis in pMMR CRC and increased production of amino acid proxies for hydrogen sulfide in dMMR CRC.
Conclusions: Integrating tumor biology and microbial ecology highlighted distinct microbial, metabolic, and ecological properties unique to dMMR and pMMR CRC. This approach could critically improve our ability to define, predict, prevent, and treat colorectal cancers.
Conflict of interest statement
Ethics approval and consent to participate
This study was performed with the approval of the Mayo Clinic Institutional Review Board (IRB# 14-007237 and IRB# 622-00) in accordance with the principles of the Declaration of Helsinki. Written informed consent was obtained from all individuals in the study.
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

