Characterization of Squamous Cell Lung Cancers from Appalachian Kentucky
- PMID: 30377206
- PMCID: PMC6363884
- DOI: 10.1158/1055-9965.EPI-17-0984
Characterization of Squamous Cell Lung Cancers from Appalachian Kentucky
Abstract
Background: Lung cancer is the leading cause of cancer mortality in the United States (U.S.). Squamous cell carcinoma (SQCC) represents 22.6% of all lung cancers nationally, and 26.4% in Appalachian Kentucky (AppKY), where death from lung cancer is exceptionally high. The Cancer Genome Atlas (TCGA) characterized genetic alterations in lung SQCC, but this cohort did not focus on AppKY residents.
Methods: Whole-exome sequencing was performed on tumor and normal DNA samples from 51 lung SQCC subjects from AppKY. Somatic genomic alterations were compared between the AppKY and TCGA SQCC cohorts.
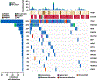
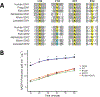
Results: From this AppKY cohort, we identified an average of 237 nonsilent mutations per patient and, in comparison with TCGA, we found that PCMTD1 (18%) and IDH1 (12%) were more commonly altered in AppKY versus TCGA. Using IDH1 as a starting point, we identified a mutually exclusive mutational pattern (IDH1, KDM6A, KDM4E, JMJD1C) involving functionally related genes. We also found actionable mutations (10%) and/or intermediate or high-tumor mutation burden (65%), indicating potential therapeutic targets in 65% of subjects.
Conclusions: This study has identified an increased percentage of IDH1 and PCMTD1 mutations in SQCC arising in the AppKY residents versus TCGA, with population-specific implications for the personalized treatment of this disease.
Impact: Our study is the first report to characterize genomic alterations in lung SQCC from AppKY. These findings suggest population differences in the genetics of lung SQCC between AppKY and U.S. populations, highlighting the importance of the relevant population when developing personalized treatment approaches for this disease.
©2018 American Association for Cancer Research.
Conflict of interest statement
Figures
Similar articles
-
Characterization of Uterine Cervix Cancers in Women from Appalachian Kentucky.Front Oncol. 2021 Dec 9;11:808081. doi: 10.3389/fonc.2021.808081. eCollection 2021. Front Oncol. 2021. PMID: 34956914 Free PMC article.
-
Genetic alteration and PD-L1 expression profiles of Chinese patients with lung squamous cell carcinoma.Pathol Res Pract. 2022 Mar;231:153761. doi: 10.1016/j.prp.2022.153761. Epub 2022 Jan 8. Pathol Res Pract. 2022. PMID: 35151031
-
Personalized therapy on the horizon for squamous cell carcinoma of the lung.Lung Cancer. 2013 Jun;80(3):249-55. doi: 10.1016/j.lungcan.2013.02.015. Epub 2013 Mar 13. Lung Cancer. 2013. PMID: 23489560 Review.
-
Squamous Cell Carcinoma in Never Smokers: An Insight into SMARCB1 Loss.Int J Mol Sci. 2024 Jul 26;25(15):8165. doi: 10.3390/ijms25158165. Int J Mol Sci. 2024. PMID: 39125735 Free PMC article.
-
Genomic landscape of squamous cell carcinoma of the lung.Am Soc Clin Oncol Educ Book. 2013:348-53. doi: 10.14694/EdBook_AM.2013.33.348. Am Soc Clin Oncol Educ Book. 2013. PMID: 23714544 Review.
Cited by
-
Cancer Informatics for Cancer Centers: Sharing Ideas on How to Build an Artificial Intelligence-Ready Informatics Ecosystem for Radiation Oncology.JCO Clin Cancer Inform. 2023 Sep;7:e2300136. doi: 10.1200/CCI.23.00136. JCO Clin Cancer Inform. 2023. PMID: 38055914 Free PMC article.
-
A probabilistic method for leveraging functional annotations to enhance estimation of the temporal order of pathway mutations during carcinogenesis.BMC Bioinformatics. 2019 Dec 2;20(1):620. doi: 10.1186/s12859-019-3218-2. BMC Bioinformatics. 2019. PMID: 31791231 Free PMC article.
-
Exceptional Survival Among Kentucky Stage IV Non-small Cell Lung Cancer Patients: Appalachian Versus Non-Appalachian Populations.J Rural Health. 2022 Jan;38(1):14-27. doi: 10.1111/jrh.12537. Epub 2020 Nov 18. J Rural Health. 2022. PMID: 33210370 Free PMC article.
-
Description of a Lung Cancer Hotspot: Disparities in Lung Cancer Histology, Incidence, and Survival in Kentucky and Appalachian Kentucky.Clin Lung Cancer. 2021 Nov;22(6):e911-e920. doi: 10.1016/j.cllc.2021.03.007. Epub 2021 Mar 26. Clin Lung Cancer. 2021. PMID: 33958300 Free PMC article.
-
SYNE1 Mutation Is Associated with Increased Tumor Mutation Burden and Immune Cell Infiltration in Ovarian Cancer.Int J Mol Sci. 2023 Sep 18;24(18):14212. doi: 10.3390/ijms241814212. Int J Mol Sci. 2023. PMID: 37762518 Free PMC article.
References
-
- Howlader N et al. SEER Cancer Statistics Review, 1975–2012, <https://seer.cancer.gov/index.html> (2015).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

