S-Locus F-Box Proteins Are Solely Responsible for S-RNase-Based Self-Incompatibility of Petunia Pollen
- PMID: 30377238
- PMCID: PMC6354264
- DOI: 10.1105/tpc.18.00615
S-Locus F-Box Proteins Are Solely Responsible for S-RNase-Based Self-Incompatibility of Petunia Pollen
Abstract
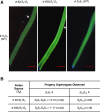
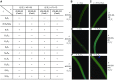
Self-incompatibility (SI) in Petunia is regulated by a polymorphic S-locus. For each S-haplotype, the S-locus contains a pistil-specific S-RNase gene and multiple pollen-specific S-locus F-box (SLF) genes. Both gain-of-function and loss-of-function experiments have shown that S-RNase alone regulates pistil specificity in SI. Gain-of-function experiments on SLF genes suggest that the entire suite of encoded proteins constitute the pollen specificity determinant. However, clear-cut loss-of-function experiments must be performed to determine if SLF proteins are essential for SI of pollen. Here, we used CRISPR/Cas9 to generate two frame-shift indel alleles of S2 -SLF1 (SLF1 of S2 -haplotype) in S2S3 plants of P. inflata and examined the effect on the SI behavior of S2 pollen. In the absence of a functional S2-SLF1, S2 pollen was either rejected by or remained compatible with pistils carrying one of eight normally compatible S-haplotypes. All results are consistent with interaction relationships between the 17 SLF proteins of S2 -haplotype and these eight S-RNases that had been determined by gain-of-function experiments performed previously or in this work. Our loss-of-function results provide definitive evidence that SLF proteins are solely responsible for SI of pollen, and they reveal their diverse and complex interaction relationships with S-RNases to maintain SI while ensuring cross-compatibility.
© 2018 American Society of Plant Biologists. All rights reserved.
Figures






Comment in
-
Self Control: SLF Proteins Are Essential for Preventing Self-Fertilization in Petunia.Plant Cell. 2018 Dec;30(12):2892-2893. doi: 10.1105/tpc.18.00828. Epub 2018 Nov 1. Plant Cell. 2018. PMID: 30389754 Free PMC article. No abstract available.
Similar articles
-
CRISPR/Cas9-mediated knockout of PiSSK1 reveals essential role of S-locus F-box protein-containing SCF complexes in recognition of non-self S-RNases during cross-compatible pollination in self-incompatible Petunia inflata.Plant Reprod. 2018 Jun;31(2):129-143. doi: 10.1007/s00497-017-0314-1. Epub 2017 Nov 30. Plant Reprod. 2018. PMID: 29192328
-
Self-incompatibility in Petunia inflata: the relationship between a self-incompatibility locus F-box protein and its non-self S-RNases.Plant Cell. 2013 Feb;25(2):470-85. doi: 10.1105/tpc.112.106294. Epub 2013 Feb 26. Plant Cell. 2013. PMID: 23444333 Free PMC article.
-
Transcriptome analysis reveals the same 17 S-locus F-box genes in two haplotypes of the self-incompatibility locus of Petunia inflata.Plant Cell. 2014 Jul;26(7):2873-88. doi: 10.1105/tpc.114.126920. Epub 2014 Jul 28. Plant Cell. 2014. PMID: 25070642 Free PMC article.
-
Self-incompatibility in Petunia: a self/nonself-recognition mechanism employing S-locus F-box proteins and S-RNase to prevent inbreeding.Wiley Interdiscip Rev Dev Biol. 2012 Mar-Apr;1(2):267-75. doi: 10.1002/wdev.10. Epub 2011 Nov 17. Wiley Interdiscip Rev Dev Biol. 2012. PMID: 23801440 Review.
-
S-RNase-based self-incompatibility in Petunia inflata.Ann Bot. 2011 Sep;108(4):637-46. doi: 10.1093/aob/mcq253. Epub 2010 Dec 30. Ann Bot. 2011. PMID: 21193481 Free PMC article. Review.
Cited by
-
Global transcriptome dissection of pollen-pistil interactions induced self-incompatibility in dragon fruit (Selenicereus spp.).PeerJ. 2022 Nov 1;10:e14165. doi: 10.7717/peerj.14165. eCollection 2022. PeerJ. 2022. PMID: 36340195 Free PMC article.
-
Ubiquitination of S4-RNase by S-LOCUS F-BOX LIKE2 Contributes to Self-Compatibility of Sweet Cherry 'Lapins'.Plant Physiol. 2020 Dec;184(4):1702-1716. doi: 10.1104/pp.20.01171. Epub 2020 Oct 9. Plant Physiol. 2020. PMID: 33037127 Free PMC article.
-
Diverse Evolution in 111 Plant Genomes Reveals Purifying and Dosage Balancing Selection Models for F-Box Genes.Int J Mol Sci. 2021 Jan 16;22(2):871. doi: 10.3390/ijms22020871. Int J Mol Sci. 2021. PMID: 33467195 Free PMC article.
-
Self Control: SLF Proteins Are Essential for Preventing Self-Fertilization in Petunia.Plant Cell. 2018 Dec;30(12):2892-2893. doi: 10.1105/tpc.18.00828. Epub 2018 Nov 1. Plant Cell. 2018. PMID: 30389754 Free PMC article. No abstract available.
-
Transcriptome Analysis of the Late-Acting Self-Incompatibility Associated with RNase T2 Family in Camellia oleifera.Plants (Basel). 2023 May 9;12(10):1932. doi: 10.3390/plants12101932. Plants (Basel). 2023. PMID: 37653852 Free PMC article.
References
-
- Ai Y., Singh A., Coleman C.E., Ioerger T.R., Kheyr-Pour A., Kao T.H. (1990). Self-incompatibility in Petunia inflata: Isolation and characterization of cDNAs encoding three S-allele-associated proteins. Sex. Plant Reprod. 3: 130–138.
-
- Bombarely A., et al. (2016). Insight into the evolution of the Solanaceae from the parental genomes of Petunia hybrida. Nat. Plants 2: 16074. - PubMed
-
- de Nettancourt D. (2001). Incompatibility and Incongruity in Wild and Cultivated Plants. (Berlin, NY: Springer; ).
-
- Entani T., Kubo K., Isogai S., Fukao Y., Shirakawa M., Isogai A., Takayama S. (2014). Ubiquitin-proteasome-mediated degradation of S-RNase in a solanaceous cross-compatibility reaction. Plant J. 78: 1014–1021. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

