KDM3A histone demethylase functions as an essential factor for activation of JAK2-STAT3 signaling pathway
- PMID: 30377265
- PMCID: PMC6243239
- DOI: 10.1073/pnas.1805662115
KDM3A histone demethylase functions as an essential factor for activation of JAK2-STAT3 signaling pathway
Abstract
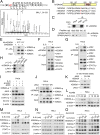
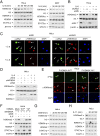
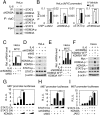
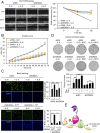
Janus tyrosine kinase 2 (JAK2)-signal transducer and activator of transcription 3 (STAT3) signaling pathway is essential for modulating cellular development, differentiation, and homeostasis. Thus, dysregulation of JAK2-STAT3 signaling pathway is frequently associated with human malignancies. Here, we provide evidence that lysine-specific demethylase 3A (KDM3A) functions as an essential epigenetic enzyme for the activation of JAK2-STAT3 signaling pathway. KDM3A is tyrosine-phosphorylated by JAK2 in the nucleus and functions as a STAT3-dependent transcriptional coactivator. JAK2-KDM3A signaling cascade induced by IL-6 leads to alteration of histone H3K9 methylation as a predominant epigenetic event, thereby providing the functional and mechanistic link between activation of JAK2-STAT3 signaling pathway and its epigenetic control. Together, our findings demonstrate that inhibition of KDM3A phosphorylation could be a potent therapeutic strategy to control oncogenic effect of JAK2-STAT3 signaling pathway.
Keywords: JAK2; KDM3A; STAT3; histone demethylation; phosphorylation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
The histone demethylase KDM3A regulates the transcriptional program of the androgen receptor in prostate cancer cells.Oncotarget. 2017 May 2;8(18):30328-30343. doi: 10.18632/oncotarget.15681. Oncotarget. 2017. PMID: 28416760 Free PMC article.
-
[H3K27me3 Expression Level and Its Epigenetic Regulation Mechanisms in Th17 Cell Differentiation in Patients With Ankylosing Spondylitis].Sichuan Da Xue Xue Bao Yi Xue Ban. 2024 May 20;55(3):744-748. doi: 10.12182/20240560605. Sichuan Da Xue Xue Bao Yi Xue Ban. 2024. PMID: 38948276 Free PMC article. Chinese.
-
Impact of histone demethylase KDM3A-dependent AP-1 transactivity on hepatotumorigenesis induced by PI3K activation.Oncogene. 2017 Nov 9;36(45):6262-6271. doi: 10.1038/onc.2017.222. Epub 2017 Jul 10. Oncogene. 2017. PMID: 28692045
-
Epigenetic gene regulation by plant Jumonji group of histone demethylase.Biochim Biophys Acta. 2011 Aug;1809(8):421-6. doi: 10.1016/j.bbagrm.2011.03.004. Epub 2011 Mar 16. Biochim Biophys Acta. 2011. PMID: 21419882 Review.
-
The bidirectional role of the JAK2/STAT3 signaling pathway and related mechanisms in cerebral ischemia-reperfusion injury.Exp Neurol. 2021 Jul;341:113690. doi: 10.1016/j.expneurol.2021.113690. Epub 2021 Mar 31. Exp Neurol. 2021. PMID: 33798563 Review.
Cited by
-
FGL1: a novel biomarker and target for non-small cell lung cancer, promoting tumor progression and metastasis through KDM4A/STAT3 transcription mechanism.J Exp Clin Cancer Res. 2024 Aug 1;43(1):213. doi: 10.1186/s13046-024-03140-6. J Exp Clin Cancer Res. 2024. PMID: 39085849 Free PMC article.
-
Post-translational modification analysis of Saccharomyces cerevisiae histone methylation enzymes reveals phosphorylation sites of regulatory potential.J Biol Chem. 2021 Jan-Jun;296:100192. doi: 10.1074/jbc.RA120.015995. Epub 2020 Dec 20. J Biol Chem. 2021. PMID: 33334889 Free PMC article.
-
The Anti-Proliferative Activity of the Hybrid TMS-TMF-4f Compound Against Human Cervical Cancer Involves Apoptosis Mediated by STAT3 Inactivation.Cancers (Basel). 2019 Dec 3;11(12):1927. doi: 10.3390/cancers11121927. Cancers (Basel). 2019. PMID: 31816985 Free PMC article.
-
Expression of proliferation-related genes in BM-MSC-treated ALL cells in hypoxia condition is regulated under the influence of epigenetic factors in-vitro.Med Oncol. 2022 May 18;39(5):88. doi: 10.1007/s12032-022-01671-6. Med Oncol. 2022. PMID: 35581482
-
The evolving landscape of epigenetic target molecules and therapies in myeloid cancers: focus on acute myeloid leukemia and myeloproliferative neoplasms.Leukemia. 2025 Aug;39(8):1824-1837. doi: 10.1038/s41375-025-02639-x. Epub 2025 May 15. Leukemia. 2025. PMID: 40374809 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

