Nuclear envelope expansion in budding yeast is independent of cell growth and does not determine nuclear volume
- PMID: 30379612
- PMCID: PMC6337908
- DOI: 10.1091/mbc.E18-04-0204
Nuclear envelope expansion in budding yeast is independent of cell growth and does not determine nuclear volume
Abstract
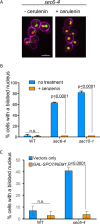
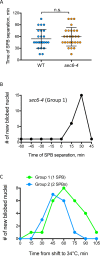
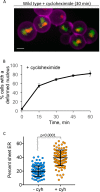
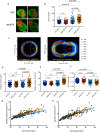
Most cells exhibit a constant ratio between nuclear and cell volume. The mechanism dictating this constant ratio and the nuclear component(s) that scale with cell size are not known. To address this, we examined the consequences to the size and shape of the budding yeast nucleus when cell expansion is inhibited by down-regulating components of the secretory pathway. We find that under conditions where cell size increase is restrained, the nucleus becomes bilobed, with the bulk of the DNA in one lobe and the nucleolus in the other. The formation of bilobed nuclei is dependent on fatty acid and phospholipid synthesis, suggesting that it is associated with nuclear membrane expansion. Bilobed nuclei appeared predominantly after spindle pole body separation, suggesting that nuclear envelope expansion follows cell-cycle cues rather than cell size. Importantly, cells with bilobed nuclei had the same nuclear:cell volume ratio as cells with round nuclei. Therefore, the bilobed nucleus could be a consequence of continued NE expansion as cells traverse the cell cycle without an accompanying increase in nuclear volume due to the inhibition of cell growth. Our data suggest that nuclear volume is not determined by nuclear envelope availability but by one or more nucleoplasmic factors.
Figures





References
-
- Berger AB, Cabal GG, Fabre E, Duong T, Buc H, Nehrbass U, Olivo-Marin J-C, Gadal O, Zimmer C. (2008). High-resolution statistical mapping reveals gene territories in live yeast. Nat Methods , 1031–1037. - PubMed
-
- Blumberg H, Silver PA. (1991). A split zinc-finger protein is required for normal yeast growth. Gene , 101–110. - PubMed

