Identification of Plasmodium falciparum nuclear proteins by mass spectrometry and proposed protein annotation
- PMID: 30379851
- PMCID: PMC6209197
- DOI: 10.1371/journal.pone.0205596
Identification of Plasmodium falciparum nuclear proteins by mass spectrometry and proposed protein annotation
Abstract
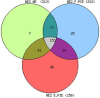
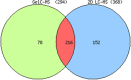
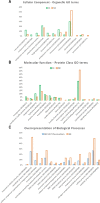
The nuclear proteome of Plasmodium falciparum results from the continual shuttle of proteins between the cell cytoplasm-nucleus and vice versa. Using shotgun proteomics tools, we explored the nuclear proteins of mixed populations of Plasmodium falciparum extracted from infected erythrocytes. We combined GeLC-MS/MS and 2D-LC-MS/MS with a peptide ion exclusion procedure in order to increase the detection of low abundant proteins such as those involved in gene expression. We have identified 446 nuclear proteins covering all expected nuclear protein families involved in gene regulation. All structural ribosomal (40S and 60S) proteins were identified which is consistent with the nuclear localization of ribosomal biogenesis. Proteins involved in the translation machinery were also found suggesting that translational events might occur in the nucleus in P. falciparum as previously hypothesized in eukaryotes. These data were compared to the protein list established by PlasmoDB and submitted to Plasmobase a recently reported Plasmodium annotation website to propose new functional putative annotation of several unknown proteins found in the nuclear extracts.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

