Harmonization of exosome isolation from culture supernatants for optimized proteomics analysis
- PMID: 30379855
- PMCID: PMC6209201
- DOI: 10.1371/journal.pone.0205496
Harmonization of exosome isolation from culture supernatants for optimized proteomics analysis
Abstract
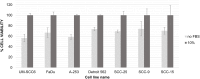
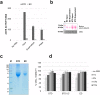
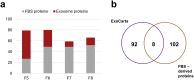
Exosomes, the smallest subset of extracellular vesicles (EVs), have recently attracted much attention in the scientific community. Their involvement in intercellular communication and molecular reprogramming of different cell types created a demand for a stringent characterization of the proteome which exosomes carry and deliver to recipient cells. Mass spectrometry (MS) has been extensively used for exosome protein profiling. Unfortunately, no standards have been established for exosome isolation and their preparation for MS, leading to accumulation of artefactual data. These include the presence of high-abundance exosome-contaminating serum proteins in culture media which mask low-abundance exosome-specific components, isolation methods that fail to yield "pure" vesicles or variability in protein solubilization protocols. There is an unmet need for the development of standards for exosome generation, harvesting, and isolation from cellular supernatants and for optimization of protein extraction methods before proteomics analysis by MS. In this communication, we illustrate the existing problems in this field and provide a set of recommendations that are expected to harmonize exosome processing for MS and provide the faithful picture of the proteomes carried by exosomes. The recommended workflow for effective and specific identification of proteins in exosomes released by the low number of cells involves culturing cells in medium with a reduced concentration of exosome-depleted serum, purification of exosomes by size-exclusion chromatography, a combination of different protein extraction method and removal of serum-derived proteins from the final dataset using an appropriate sample of cell-unexposed medium as a control. Application of this method allowed detection of >250 vesicle-specific proteins in exosomes from 10 mL of culture medium.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

